1-DAV-202 Data Management 2024/25
Difference between revisions of "Lbioinf1"
Jump to navigation
Jump to search
| (7 intermediate revisions by the same user not shown) | |||
| Line 6: | Line 6: | ||
==Overview of DNA sequencing and assembly== | ==Overview of DNA sequencing and assembly== | ||
| − | '''DNA sequencing''' is a technology of reading the order of nucleotides along a DNA strand | + | '''DNA sequencing''' is a technology of reading the order of nucleotides along a DNA strand. |
| − | * The result is represented as a string of A,C,G,T | + | * The result is represented as a string of A,C,G,T. |
| − | * Only fragments of DNA of limited length can be read, these are called '''sequencing reads''' | + | * Only fragments of DNA of limited length can be read, these are called '''sequencing reads'''. |
| − | * Different technologies produce reads of different characteristics | + | * Different technologies produce reads of different characteristics. |
* Examples: | * Examples: | ||
| − | ** '''Illumina sequencers''' produce short reads (typical length 100-200bp), but in great quantities and very low error rate (<0.1%) | + | ** '''Illumina sequencers''' produce short reads (typical length 100-200bp), but in great quantities and very low error rate (<0.1%). |
| − | ** Illumina reads usually come in '''pairs''' sequenced from both ends of a DNA fragment of an approximately known length | + | ** Illumina reads usually come in '''pairs''' sequenced from both ends of a DNA fragment of an approximately known length. |
| − | ** '''Oxford nanopore sequencers''' produce longer reads (thousands of | + | ** '''Oxford nanopore sequencers''' produce longer reads (thousands of base pairs or more), but the error rates are higher (2-15%). |
| − | The goal of '''genome sequencing''' is to read all chromosomes of an organism | + | The goal of '''genome sequencing''' is to read all chromosomes of an organism. |
| − | * Sequencing machines produce many reads coming from different parts of the genome | + | * Sequencing machines produce many reads coming from different parts of the genome. |
| − | * Using software tools called '''sequence assemblers''', these reads are glued together based on overlaps | + | * Using software tools called '''sequence assemblers''', these reads are glued together based on overlaps. |
| − | * Ideally we would get the true chromosomes, but often we get only shorter fragments called '''contigs''' | + | * Ideally we would get the true chromosomes, but often we get only shorter fragments called '''contigs'''. |
| − | * | + | * Assembled contigs sometimes contain errors. |
| − | * We prefer longer contigs with fewer errors | + | * We prefer longer contigs with fewer errors. |
==Sequence alignments and dotplots== | ==Sequence alignments and dotplots== | ||
| − | * '''Sequence alignment''' is the task of finding similarities between DNA (or protein) sequences | + | <!-- NOTEX --> |
| − | * Here is an example | + | A short video for this section: [https://youtu.be/qANrSl5w4t8] |
| + | <!-- /NOTEX --> | ||
| + | * '''Sequence alignment''' is the task of finding similarities between DNA (or protein) sequences. | ||
| + | * Here is an example, a short similarity between region at positions 344,447..344,517 of one sequence and positions 3,261..3,327 of another sequence. | ||
<pre> | <pre> | ||
Query: 344447 tctccgacggtgatggcgttgtgcgtcctctatttcttttatttctttttgttttatttc 344506 | Query: 344447 tctccgacggtgatggcgttgtgcgtcctctatttcttttatttctttttgttttatttc 344506 | ||
| Line 36: | Line 39: | ||
</pre> | </pre> | ||
| − | * Alignments can be stored in many formats and | + | * Alignments can be stored in many formats and visualized as dotplots. |
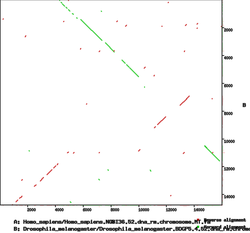
| − | * In a '''dotplot''', the x-axis correspond to positions in one sequence and the y-axis in another sequence | + | * In a '''dotplot''', the x-axis correspond to positions in one sequence and the y-axis in another sequence. |
| − | * Diagonal lines show alignments between the sequences (direction of the diagonal shows which DNA strand was aligned) | + | * Diagonal lines show alignments between the sequences (direction of the diagonal shows which DNA strand was aligned). |
[[Image:Dotplot-mt-human-dros.png|center|thumb|250px|Dotplot of human and ''Drosophila'' mitochondrial genomes]] | [[Image:Dotplot-mt-human-dros.png|center|thumb|250px|Dotplot of human and ''Drosophila'' mitochondrial genomes]] | ||
| Line 45: | Line 48: | ||
===FASTA=== | ===FASTA=== | ||
| − | * FASTA is a format for storing DNA, RNA and protein sequences | + | * FASTA is a format for storing DNA, RNA and protein sequences. |
| − | * We have already seen FASTA files in [[HWperl|Perl exercises]] | + | * We have already seen FASTA files in [[HWperl|Perl exercises]]. |
| − | * Each sequence is given on several lines of the file. The first line starts with ">" followed by an identifier of the sequence and optionally some further description separated by whitespace | + | * Each sequence is given on several lines of the file. The first line starts with ">" followed by an identifier of the sequence and optionally some further description separated by whitespace. |
| − | * The sequence itself is on the second line; long sequences are split into multiple lines | + | * The sequence itself is on the second line; long sequences are split into multiple lines. |
<pre> | <pre> | ||
>SRR022868.1845_1 | >SRR022868.1845_1 | ||
| − | + | AAATTTAGGAAAAGATGATTTAGCAACATTTAGCCTTAATGAAAGACCAGATTCTGTTGCCATGTTTGAA... | |
>SRR022868.1846_1 | >SRR022868.1846_1 | ||
| − | + | TAGCGTTGTAAAATAAATTTCTAGAATGGAAGTGATGATATTGAAATACACTCAGATCCTGAATGAAAGA... | |
</pre> | </pre> | ||
===FASTQ=== | ===FASTQ=== | ||
| − | * FASTQ is a format for storing sequencing reads, containing DNA sequences but also quality information about each nucleotide | + | * FASTQ is a format for storing sequencing reads, containing DNA sequences but also quality information about each nucleotide. |
| − | * More in the [[Lperl#The_second_input_file_for_today:_DNA_sequencing_reads_.28fastq.29|lecture on Perl]] | + | * More in the [[Lperl#The_second_input_file_for_today:_DNA_sequencing_reads_.28fastq.29|lecture on Perl]]. |
===SAM/BAM=== | ===SAM/BAM=== | ||
| − | * SAM and BAM are formats for storing alignments of sequencing reads (or other sequences) to a genome | + | * SAM and BAM are formats for storing alignments of sequencing reads (or other sequences) to a genome. |
| − | * For each read, the file contains the read itself, its quality, but also the chromosome/contig name and position where this read is likely coming from, and an additional information e.g. about mapping quality (confidence in the correct location) | + | * For each read, the file contains the read itself, its quality, but also the chromosome/contig name and position where this read is likely coming from, and an additional information e.g. about mapping quality (confidence in the correct location). |
| − | * SAM files are text-based, thus easier to check manually; BAM files are binary and compressed, thus smaller and faster to read | + | * SAM files are text-based, thus easier to check manually; BAM files are binary and compressed, thus smaller and faster to read. |
| − | * We can easily convert between SAM and BAM using [https://github.com/samtools/samtools samtools] | + | * We can easily convert between SAM and BAM using [https://github.com/samtools/samtools samtools]. |
* [https://samtools.github.io/hts-specs/SAMv1.pdf Full documentation of the format] | * [https://samtools.github.io/hts-specs/SAMv1.pdf Full documentation of the format] | ||
===PAF format=== | ===PAF format=== | ||
| − | * PAF is another format for storing alignments used in the minimap2 tool | + | * PAF is another format for storing alignments used in the minimap2 tool. |
* [https://github.com/lh3/miniasm/blob/master/PAF.md Full documentation of the format] | * [https://github.com/lh3/miniasm/blob/master/PAF.md Full documentation of the format] | ||
===Gzip=== | ===Gzip=== | ||
| − | * Gzip is a general-purpose tool for file compression | + | * Gzip is a general-purpose tool for file compression. |
| − | * It is often used in bioinformatics on large FASTQ or FASTA files | + | * It is often used in bioinformatics on large FASTQ or FASTA files. |
| − | * Running command <tt>gzip filename.ext</tt> will create compressed file <tt>filename.ext.gz</tt> | + | * Running command <tt>gzip filename.ext</tt> will create compressed file <tt>filename.ext.gz</tt> and the original file will be deleted. |
| − | * The reverse process is done by <tt>gunzip filename.ext.gz</tt> | + | * The reverse process is done by <tt>gunzip filename.ext.gz</tt>. This deletes the gziped file and creates the uncompressed version. |
| − | * However, we can access the file | + | * However, we can access the file without uncompressing it. Command <tt>zcat filename.ext.gz</tt> prints the content of a gzipped file and keeps the gzipped file as is. We can use pipes <tt>|</tt> to do further processing on the file. |
| − | * To manually page through the content of a gzipped file use <tt>zless filename.ext.gz</tt> | + | * To manually page through the content of a gzipped file use <tt>zless filename.ext.gz</tt>. |
| − | * Some bioinformatics tools can work directly with gzipped files | + | * Some bioinformatics tools can work directly with gzipped files. |
Latest revision as of 12:31, 13 March 2023
The next three lectures at targeted at the students in the Bioinformatics program and the goal is to get experience with several common bioinformatics tools. Students will learn more about the algorithms and models behind these tools in the Methods in bioinformatics course.
Contents
Overview of DNA sequencing and assembly
DNA sequencing is a technology of reading the order of nucleotides along a DNA strand.
- The result is represented as a string of A,C,G,T.
- Only fragments of DNA of limited length can be read, these are called sequencing reads.
- Different technologies produce reads of different characteristics.
- Examples:
- Illumina sequencers produce short reads (typical length 100-200bp), but in great quantities and very low error rate (<0.1%).
- Illumina reads usually come in pairs sequenced from both ends of a DNA fragment of an approximately known length.
- Oxford nanopore sequencers produce longer reads (thousands of base pairs or more), but the error rates are higher (2-15%).
The goal of genome sequencing is to read all chromosomes of an organism.
- Sequencing machines produce many reads coming from different parts of the genome.
- Using software tools called sequence assemblers, these reads are glued together based on overlaps.
- Ideally we would get the true chromosomes, but often we get only shorter fragments called contigs.
- Assembled contigs sometimes contain errors.
- We prefer longer contigs with fewer errors.
Sequence alignments and dotplots
A short video for this section: [1]
- Sequence alignment is the task of finding similarities between DNA (or protein) sequences.
- Here is an example, a short similarity between region at positions 344,447..344,517 of one sequence and positions 3,261..3,327 of another sequence.
Query: 344447 tctccgacggtgatggcgttgtgcgtcctctatttcttttatttctttttgttttatttc 344506
|||||||| |||||||||||||||||| ||||||| |||||||||||| || ||||||
Sbjct: 3261 tctccgacagtgatggcgttgtgcgtc-tctatttattttatttctttgtg---tatttc 3316
Query: 344507 tctgactaccg 344517
|||||||||||
Sbjct: 3317 tctgactaccg 3327
- Alignments can be stored in many formats and visualized as dotplots.
- In a dotplot, the x-axis correspond to positions in one sequence and the y-axis in another sequence.
- Diagonal lines show alignments between the sequences (direction of the diagonal shows which DNA strand was aligned).
File formats
FASTA
- FASTA is a format for storing DNA, RNA and protein sequences.
- We have already seen FASTA files in Perl exercises.
- Each sequence is given on several lines of the file. The first line starts with ">" followed by an identifier of the sequence and optionally some further description separated by whitespace.
- The sequence itself is on the second line; long sequences are split into multiple lines.
>SRR022868.1845_1 AAATTTAGGAAAAGATGATTTAGCAACATTTAGCCTTAATGAAAGACCAGATTCTGTTGCCATGTTTGAA... >SRR022868.1846_1 TAGCGTTGTAAAATAAATTTCTAGAATGGAAGTGATGATATTGAAATACACTCAGATCCTGAATGAAAGA...
FASTQ
- FASTQ is a format for storing sequencing reads, containing DNA sequences but also quality information about each nucleotide.
- More in the lecture on Perl.
SAM/BAM
- SAM and BAM are formats for storing alignments of sequencing reads (or other sequences) to a genome.
- For each read, the file contains the read itself, its quality, but also the chromosome/contig name and position where this read is likely coming from, and an additional information e.g. about mapping quality (confidence in the correct location).
- SAM files are text-based, thus easier to check manually; BAM files are binary and compressed, thus smaller and faster to read.
- We can easily convert between SAM and BAM using samtools.
- Full documentation of the format
PAF format
- PAF is another format for storing alignments used in the minimap2 tool.
- Full documentation of the format
Gzip
- Gzip is a general-purpose tool for file compression.
- It is often used in bioinformatics on large FASTQ or FASTA files.
- Running command gzip filename.ext will create compressed file filename.ext.gz and the original file will be deleted.
- The reverse process is done by gunzip filename.ext.gz. This deletes the gziped file and creates the uncompressed version.
- However, we can access the file without uncompressing it. Command zcat filename.ext.gz prints the content of a gzipped file and keeps the gzipped file as is. We can use pipes | to do further processing on the file.
- To manually page through the content of a gzipped file use zless filename.ext.gz.
- Some bioinformatics tools can work directly with gzipped files.