1-DAV-202 Data Management 2024/25
Lbioinf2
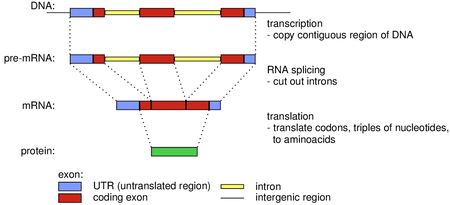
Eukaryotic gene structure
- Recall the Central dogma of molecular biology: the flow of genetic information from DNA to RNA to protein (gene expression)
- In eukaryotes, mRNA often undergoes splicing, where introns are removed and exons are joined together
- The very start and end of mRNA remain untranslated (UTR = untranslated region)
- The coding part of the gene starts with a start codon, contains a sequence of additional codons and ends with a stop codon. Codons can be interrupted by introns.
Computational gene finding
- Input: DNA sequence (an assembled genome or a part of it)
- Output: positions of protein coding genes and their exons
- If we know the exact position of coding regions of a gene, we can use the genetic code table to predict the protein sequence encoded by it.
- Gene finders use statistical features observed from known genes, such as typical sequence motifs near the start codons, stop codons and splice sites, typical codon frequencies, typical exon and intron lengths etc.
- These statistical parameters need to be adjusted for each genome.
- We will use a gene finder called Augustus.
Gene expression
- Not all genes undergo transcription and translation all the time and at the same level.
- The processes of transcription and translation are regulated according to cell needs.
- The term "gene expression" has two meanings:
- the process of transcription and translation (synthesis of a gene product),
- the amount of mRNA or protein produced from a single gene (genes with high or low expression).
RNA-seq technology can sequence mRNA extracted from a sample of cells.
- We can align sequenced reads back to the genome.
- The number of reads coming from a gene depends on its expression level (and on its length).
File formats
In addition to file formats from last lecture, we will today encounter several new formats:
- GTF format is used to store location of genes and their exons. Rows starting with # are comments, each of the remaining rows describes some interval of the sequence. If the second column is CDS, it is a coding part of an exon.
- GFF3 format is similar to GTF, used for the same purpose.
- BED format is used to describe location of arbitrary elements in the genome. It has variable number of columns: the first 3 columns with sequence name, start and end are compulsory, but more columns can be added with more details if available.