1-DAV-202 Data Management 2024/25
Difference between revisions of "Integrácia dátových zdrojov 2016/17"
| (One intermediate revision by the same user not shown) | |||
| Line 1: | Line 1: | ||
Website for 2017/17 | Website for 2017/17 | ||
| − | * [[Kontakt]] | + | * [[#Kontakt]] |
| − | * [[Úvod]] | + | * [[#Úvod]] |
| − | * [[Pravidlá]] | + | * [[#Pravidlá]] |
| − | * 2017-02-21 (TV) Perl, part 1 (basics, input processing): [[L01|Lecture 1]], [[HW01|Homework 1]] | + | * 2017-02-21 (TV) Perl, part 1 (basics, input processing): [[#L01|Lecture 1]], [[#HW01|Homework 1]] |
| − | * 2017-02-28 (TV) Perl, part 2 (external commands, files, subroutines): [[L02|Lecture 2]], [[HW02|Homework 2]] | + | * 2017-02-28 (TV) Perl, part 2 (external commands, files, subroutines): [[#L02|Lecture 2]], [[#HW02|Homework 2]] |
| − | * 2017-03-07 (BB) Command-line tools, Perl one-liners: [[L03|Lecture 3]], [[HW03|Homework 3]] | + | * 2017-03-07 (BB) Command-line tools, Perl one-liners: [[#L03|Lecture 3]], [[#HW03|Homework 3]] |
| − | * 2017-03-14 (BB) Job scheduling and make [[L04|Lecture 4]], [[HW04|Homework 4]] | + | * 2017-03-14 (BB) Job scheduling and make [[#L04|Lecture 4]], [[#HW04|Homework 4]] |
| − | * 2017-03-21 (BB) R, part 1 [[L05|Lecture 5]], [[HW05|Homework 5]] | + | * 2017-03-21 (BB) R, part 1 [[#L05|Lecture 5]], [[#HW05|Homework 5]] |
| − | * 2017-03-28 (BB) R, part 2 [[L06|Lecture 6]], [[HW06|Homework 6]] | + | * 2017-03-28 (BB) R, part 2 [[#L06|Lecture 6]], [[#HW06|Homework 6]] |
| − | * 2017-04-04 (VB) Python, web crawling, HTML parsing, sqlite3 [[L07|Lecture 7]], [[HW07|Homework 7]] | + | * 2017-04-04 (VB) Python, web crawling, HTML parsing, sqlite3 [[#L07|Lecture 7]], [[#HW07|Homework 7]] |
| − | * 2017-04-11 Python and SQL for beginners (work on | + | * 2017-04-11 Python and SQL for beginners (work on HW07), consultations on project topics |
* 2017-04-18 no lecture (Easter) | * 2017-04-18 no lecture (Easter) | ||
| − | * 2017-04-25 (VB) Text data processing, flask [[L08|Lecture 8]], [[HW08|Homework 8]] | + | * 2017-04-25 (VB) Text data processing, flask [[#L08|Lecture 8]], [[#HW08|Homework 8]] |
| − | * 2017-05-02 (VB) Data visualization in JavaScript [[L09|Lecture 9]], [[HW09|Homework 9]] | + | * 2017-05-02 (VB) Data visualization in JavaScript [[#L09|Lecture 9]], [[#HW09|Homework 9]] |
| − | * 2017-05-09 (JS) Hadoop [[L10|Lecture 10]], [[HW10|Homework 10]] | + | * 2017-05-09 (JS) Hadoop [[#L10|Lecture 10]], [[#HW10|Homework 10]] |
| − | * 2017-05-16 (BB) More databases, scripting language of your choice [[L11|Lecture 11]], [[HW11|Homework 11]] | + | * 2017-05-16 (BB) More databases, scripting language of your choice [[#L11|Lecture 11]], [[#HW11|Homework 11]] |
* 2017-05-23 work on projects | * 2017-05-23 work on projects | ||
| + | |||
| + | =Kontakt= | ||
| + | '''Vyučujúci''' | ||
| + | |||
| + | * [http://compbio.fmph.uniba.sk/~bbrejova/ doc. Mgr. Broňa Brejová, PhD.] miestnosť M-163 <!-- , [[Image:e-bb.png]] --> | ||
| + | * [http://compbio.fmph.uniba.sk/~tvinar/ Mgr. Tomáš Vinař, PhD.], miestnosť M-163 <!-- , [[Image:e-tv.png]] --> | ||
| + | * [http://dai.fmph.uniba.sk/w/Vladimir_Boza/sk Mgr. Vladimír Boža], miestnosť M-25 <!-- , [[Image:e-vb.png]] --> | ||
| + | * [http://dai.fmph.uniba.sk/~siska/ RNDr. Jozef Šiška, PhD.], miestnosť I-7 | ||
| + | * Konzultácie po dohode emailom | ||
| + | |||
| + | '''Rozvrh''' | ||
| + | * Utorok 14:00-16:20 M-217 | ||
| + | =Úvod= | ||
| + | ==Cieľová skupina== | ||
| + | Tento predmet je určený pre študentov 2. ročníka bakalárskeho študijného programu Bioinformatika a pre študentov bakalárskeho a magisterského študijného programu Informatika, obzvlášť ak plánujú na magisterskom štúdiu absolvovať štátnicové zameranie Bioinformatika a strojové učenie. Radi privítame aj študentov iných zameraní a študijných programov, pokiaľ majú požadované (neformálne) prerekvizity. | ||
| + | |||
| + | Predpokladáme, že študenti na tomto predmete už vedia programovať v niektorom programovacom jazyku a neboja sa učiť podľa potreby nové jazyky. Takisto predpokladáme základnú znalosť práce v Linuxe vrátane spúšťania príkazov na príkazovom riadku (mali by ste poznať aspoň základné príkazy na prácu so súbormi a adresármi ako cd, mkdir, cp, mv, rm, chmod a pod.). Hoci väčšina technológií preberaných na tomto predmete sa dá použiť na spracovanie dát z mnohých oblastí, budeme ich často ilustrovať na príkladoch z oblasti bioinformatiky. Pokúsime sa vysvetliť potrebné pojmy, ale bolo by dobre, ak by ste sa orientovali v základných pojmoch molekulárnej biológie, ako sú DNA, RNA, proteín, gén, genóm, evolúcia, fylogenetický strom a pod. Študentom zamerania Bioinformatika a strojové učenie odporúčame absolvovať najskôr Metódy v bioinformatike, až potom tento predmet. | ||
| + | |||
| + | Ak sa chcete doučiť základy používania príkazového riadku, skúste napr. tento tutoriál: http://korflab.ucdavis.edu/bootcamp.html | ||
| + | |||
| + | ==Cieľ predmetu== | ||
| + | |||
| + | Počas štúdia sa naučíte mnohé zaujímave algoritmy, modely a metódy, ktoré sa dajú použiť na spracovanie dát v bioinformatike alebo iných oblastiach. Ak však počas štúdia alebo aj neskôr v zamestnaní budete chcieť tieto metódy použiť na reálne dáta, zistíte, že väčšinou treba vynaložiť značné úsilie na samotné získanie dát, ich predspracovanie do vhodného tvaru, testovanie a porovnávanie rôznych metód alebo ich nastavení a získavanie finálnych výsledkov v tvare prehľadných tabuliek a grafov. Často je potrebné tieto činnosti veľakrát opakovať pre rôzne vstupy, rôzne nastavenia a podobne. Obzvlášť v bioinformatike je možné si nájsť zamestnanie, kde vašou hlavnou náplňou bude spracovanie dát s použitím už hotových nástrojov, prípadne doplnených menšími vlastnými programami. Na tomto predmete si ukážeme niektoré programovacie jazyky, postupy a technológie vhodné na tieto činnosti. Veľa z nich je použiteľných na dáta z rôznych oblastí, ale budeme sa venovať aj špecificky bioinformatickým nástrojom. | ||
| + | |||
| + | ==Základné princípy== | ||
| + | |||
| + | Odporúčame nasledujúci článok s dobrými radami k výpočtovým experimentom: | ||
| + | * Noble WS. [http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1000424 A quick guide to organizing computational biology projects.] PLoS Comput Biol. 2009 Jul 31;5(7):e1000424. | ||
| + | |||
| + | Niektoré dôležité zásady: | ||
| + | * Citát z článku Noble 2009: "Everything you do, you will probably have to do over again." | ||
| + | * Dobre zdokumentujte všetky kroky experimentu (čo ste robili, prečo ste to robili, čo vám vyšlo) | ||
| + | ** Ani vy sami si o pár mesiacov tieto detaily nebudete pamätať | ||
| + | * Snažte sa udržiavať logickú štruktúru adresárov a súborov | ||
| + | ** Ak však máte veľa experimentov, môže byť dostačujúce označiť ich dátumom, nevymýšľať stále nové dlhé mená | ||
| + | * Snažte sa vyhýbať manuálnym úpravám medzivýsledkov, ktoré znemožňujú jednoduché zopakovanie experimentu | ||
| + | * Snažte sa detegovať chyby v dátach | ||
| + | ** Skripty by mali skončiť s chybovou hláškou, keď niečo nejde ako by malo | ||
| + | ** V skriptoch čo najviac kontrolujte, že vstupné dáta zodpovedajú vašim predstavám (správny formát, rozumný rozsah hodnôt atď.) | ||
| + | ** Ak v skripte voláte iný program, kontrolujte jeho exit code | ||
| + | ** Tiež čo najčastejšie kontrolujte medzivýsledky výpočtu (ručným prezeraním, výpočtom rôznych štatistík a pod.), aby ste odhalili prípadné chyby v dátach alebo vo vašom kóde | ||
| + | =Pravidlá= | ||
| + | ==Známkovanie== | ||
| + | |||
| + | * Domáce úlohy: 55% | ||
| + | * Návrh projektu: 5% | ||
| + | * Projekt: 40% | ||
| + | |||
| + | Stupnica: | ||
| + | * A: 90 a viac, B:80...89, C: 70...79, D: 60...69, E: 50...59, FX: menej ako 50% | ||
| + | |||
| + | ==Formát predmetu== | ||
| + | * Každý týždeň 3 vyučovacie hodiny, z toho cca prvá je prednáška a na ďalšie dve cvičenia. Na cvičeniach samostatne riešite príklady, ktoré doma dokončíte ako domácu úlohu. | ||
| + | * Cez skúškové obdobie budete odovzdávať projekt. Po odovzdaní projektov sa bude konať ešte diskusia o projekte s vyučujúcimi, ktorá môže ovplyvniť vaše body z projektu. | ||
| + | * Budete mať konto na Linuxovom serveri určenom pre tento predmet. Toto konto používajte len na účely tohto predmetu a snažte sa server príliš svojou aktivitou nepreťažiť, aby slúžil všetkým študentom. Akékoľvek pokusy úmyselne narušiť chod servera budú považované za vážne porušenie pravidiel predmetu. | ||
| + | |||
| + | ==Domáce úlohy== | ||
| + | * Termín DÚ týkajúcej sa aktuálnej prednášky je vždy do 9:00 v deň nasledujúcej prednášky (t.j. väčšinou o necelý týždeň od zadania). | ||
| + | * Domácu úlohu odporúčame začať robiť na cvičení, kde vám môžeme prípadne poradiť. Ak máte otázky neskôr, pýtajte sa vyučujúcich emailom. | ||
| + | * Domácu úlohu môžete robiť na ľubovoľnom počítači, pokiaľ možno pod Linuxom. Odovzdaný kód alebo príkazy by však mali byť spustiteľné na serveri pre tento predmet, nepoužívajte teda špeciálny softvér alebo nastavenia vášho počítača. | ||
| + | * Domáca úloha sa odovzdáva nakopírovaním požadovaných súborov do požadovaného adresára na serveri. Konkrétne požiadavky budú spresnené v zadaní. | ||
| + | * Ak sú mená súborov špecifikované v zadaní, dodržujte ich. Ak ich vymýšľate sami, nazvite ich rozumne. V prípade potreby si spravte aj podadresáre, napr. na jednotlivé príklady. | ||
| + | * Dbajte na prehľadnosť odovzdaného zdrojového kódu (odsadzovanie, rozumné názvy premenných, podľa potreby komentáre) | ||
| + | |||
| + | ===Protokoly=== | ||
| + | * Väčšinou bude požadovanou súčasťou úlohy textový dokument nazvaný protokol. | ||
| + | * Protokol môže byť vo formáte .txt alebo .pdf a jeho meno má byť '''protocol.pdf''' alebo '''protocol.txt''' (nakopírujte ho do odovzdaného adresára) | ||
| + | * Protokol môže byť po slovensky alebo po anglicky. | ||
| + | * V prípade použitia txt formátu a diakritiky ju kódujte v UTF8, ale pre jednoduchosť môžete protokoly písať aj bez diakritiky. Ak je protocol v pdf formáte, mali by sa v ňom dať selektovať texty. | ||
| + | * Vo väčšine úloh dostanete kostru protokolu, dodržujte ju. | ||
| + | |||
| + | '''Hlavička protokolu, vyhodnotenie''' | ||
| + | * Na vrchu protokolu uveďte meno, číslo domácej úluhy a vaše vyhodnotenie toho, ako sa vám úlohu podarilo vyriešiť. Vyhodnotenie je prehľadný zoznam všetkých príkladov zo zadania, ktoré ste aspoň začali riešiť a kódov označujúcich ich stupeň dokončenia: | ||
| + | ** kód HOTOVO uveďte, ak si myslíte, že tento príklad máte úplne a správne vyriešený | ||
| + | ** kód ČASŤ uveďte, ak ste nevyriešili príklad celý a do poznámky za kód stručne uveďte, čo máte hotové a čo nie, prípadne ktorými časťami si nie ste istí. | ||
| + | ** kód MOŽNO uveďte, ak príklad máte celý, ale nie ste si istí, či správne. Opäť v poznámke uveďte, čím si nie ste istí. | ||
| + | ** kód NIČ uveďte, ak ste príklad ani nezačali riešiť | ||
| + | * Vaše vyhodnotenie je pre nás pomôckou pri bodovaní. Príklady označené HOTOVO budeme kontrolovať námatkovo, k príkladom označeným MOŽNO sa vám pokúsime dať nejakú spätnú väzbu, takisto aj k príkladom označeným ČASŤ, kde v poznámke vyjadríte, že ste mali nejaké problémy. | ||
| + | * Pri vyhodnotení sa pokúste čo najlepšie posúdiť správnosť vašich riešení, pričom kvalita vášho seba-hodnotenia môže vplývať na celkový počet bodov. | ||
| + | |||
| + | '''Obsah protokolu''' | ||
| + | * Ak nie je v zadaní určené inak, protokol by mal obsahovať nasledovné údaje: | ||
| + | ** '''Zoznam odovzdaných súborov:''' o každom súbore uveďte jeho význam a či ste ho vyrobili ručne, získali z externých zdrojov alebo vypočítali nejakým programom. Ak máte väčšie množstvo súborov so systematickým pomenovaním, stačí vysvetliť schému názvov všeobecne. Súbory, ktorých mená sú špecifikované v zadaní, nemusíte v zozname uvádzať. | ||
| + | ** '''Postupnosť všetkých spustených príkazov,''' prípadne iných krokov, ktorými ste dospeli k získaným výsledkom. Tu uvádzajte príkazy na spracovanie dát a spúšťanie vašich či iných programov. Netreba uvádzať príkazy súvisiace so samotným programovaním (spúšťanie editora, nastavenie práv na spustenie a pod.), s kopírovaním úlohy na server a pod. Uveďte aj stručné '''komentáre''', čo bolo účelom určitého príkazu alebo skupiny príkazov. | ||
| + | ** '''Zoznam zdrojov:''' (webstránky a pod., ktoré ste pri riešení úlohy použili. Nemusíte uvádzať webstránku predmetu a zdroje odporučené priamo v zadaní. | ||
| + | Celkovo by protokol mal umožniť čitateľovi zorientovať sa vo vašich súboroch a tiež v prípade záujmu vykonať rovnaké výpočty, akými ste dospeli vy k výsledku. Nemusíte písať slohy, stačia zrozumiteľné a prehľadné heslovité poznámky. | ||
| + | |||
| + | ==Projekty== | ||
| + | |||
| + | Cieľom projektu je vyskúšať si naučené zručnosti na konkrétnom projekte spracovania dát. Vašou úlohou je zohnať si dáta, tieto dáta analyzovať niektorými technikami z prednášok, prípadne aj inými technológiami a získané výsledky zobraziť v prehľadných grafoch a tabuľkách. Ideálne je, ak sa vám podarí prísť k zaujímavým alebo užitočným záverom, ale hodnotiť budeme hlavne voľbu vhodného postupu a jeho technickú náročnosť. Rozsah samotného programovania alebo analýzy dát by mal zodpovedať zhruba dvom domácim úlohám, ale celkovo bude projekt náročnejší, lebo na rozdiel od úloh nemáte postup a dáta vopred určené, ale musíte si ich vymyslieť sami a nie vždy sa prvý nápad ukáže ako správny. V projekte môžete využiť aj existujúce nástroje a knižnice, ale pokiaľ možno používajte nástroje spúšťané na príkazovom riadku. | ||
| + | |||
| + | Zhruba v dvoch tretinách semestra budete odovzdávať '''návrh projektu''' (formát txt alebo pdf, rozsah 0.5-1 strana). V tomto návrhu uveďte, aké dáta budete spracovávať, ako ich zoženiete, čo je cieľom analýzy a aké technológie plánujete použiť. Ciele a technológie môžete počas práce na projekte mierne pozmeniť podľa okolností, mali by ste však mať počiatočnú predstavu. K návrhu vám dáme spätnú väzbu, pričom v niektorých prípadoch môže byť potrebné tému mierne alebo úplne zmeniť. Za načas odovzdaný vhodný návrh projektu získate 5% z celkovej známky. Návrh odporúčame pred odovzdaním konzultovať s vyučujúcimi. | ||
| + | |||
| + | Cez skúškové obdobie bude určený termín '''odovzdania projektu'''. Podobne ako pri domácich úlohách odovzdávajte adresár s požadovanými súbormi: | ||
| + | * Vaše '''programy a súbory s dátami''' (veľmi veľké dátové súbory vynechajte) | ||
| + | * '''Protokol''' podobne ako pri domácich úlohách | ||
| + | ** formát txt alebo pdf, stručné heslovité poznámky | ||
| + | ** obsahuje zoznam súborov, podrobný postup pri analýze dát (spustené príkazy), ako aj použité zdroje (dáta, programy, dokumentácia a iná literatúra atď) | ||
| + | * '''Správu k projektu''' vo formáte pdf. Na rozdiel od menej formálneho protokolu by správu mal tvoriť súvislý text v odbornom štýle, podobne ako napr. záverečné práce. Môžete písať po slovensky alebo po anglicky, ale pokiaľ možno gramaticky správne. Správa by mala mať tieto časti: | ||
| + | ** úvod, v ktorom vysvetlíte ciele projektu, prípadne potrebné poznatky zo skúmanej oblasti a aké dáta ste mali k dispozícii | ||
| + | ** stručný popis metód, v ktorom neuvádzajte detailne jednotlivé kroky, skôr prehľad použitého prístupu a jeho zdôvodnenie | ||
| + | ** výsledky analýzy (tabuľky, grafy a pod.) a popis týchto výsledkov, prípadne aké závery sa z nich dajú spraviť (nezabudnite vysvetliť, čo znamenajú údaje v tabuľkách, osi grafov a pod.). Okrem finálnych výsledkov analýzy uveďte aj čiastkové výsledky, ktorými ste sa snažili overovať, že pôvodné dáta a jednotlivé časti vášho postupu sa správajú rozumne. | ||
| + | ** diskusiu, v ktorej uvediete, ktoré časti projektu boli náročné a na aké problémy ste narazili, kde sa vám naopak podarilo nájsť spôsob, ako problém vyriešiť jednoducho, ktoré časti projektu by ste spätne odporúčali robiť iným než vašim postupom, čo ste sa na projekte naučili a podobne | ||
| + | |||
| + | Projekty môžete robiť aj vo '''dvojici''', vtedy však vyžadujeme rozsiahlejší projekt a každý člen by mal byť primárne zodpovedný za určitú časť projektu, čo uveďte aj v správe. Dvojice odovzdávajú jednu správu, ale po odovzdaní projektu majú stretnutie s vyučujúcimi individuálne. | ||
| + | |||
| + | Ako nájsť tému projektu: | ||
| + | * Môžete spracovať nejaké dáta, ktoré potrebujete do bakalárskej alebo diplomovej práce, prípadne aj dáta, ktoré potrebujte na iný predmet (v tom prípade uveďte v správe, o aký predmet ide a takisto upovedomte aj druhého vyučujúceho, že ste použili spracovanie dát ako projekt pre tento predmet). Obzvlášť pre BIN študentov môže byť tento predmet vhodnou príležitosťou nájsť si tému bakalárskej práce a začať na nej pracovať. | ||
| + | * Môžete skúsiť zopakovať analýzu spravenú v nejakom vedeckom článku a overiť, že dostanete tie isté výsledky. Vhodné je tiež skúsiť analýzu aj mierne obmeniť (spustiť na iné dáta, zmeniť nejaké nastavenia, zostaviť aj iný typ grafu a pod.) | ||
| + | * Môžete skúsiť nájsť niekoho, kto má dáta, ktoré by potreboval spracovať, ale nevie ako na to (môže ísť o biológov, vedcov z iných oblastí, ale aj neziskové organizácie a pod.) V prípade, že takýmto spôsobom kontaktujete tretie osoby, bolo by vhodné pracovať na projekte obzvlášť zodpovedne, aby ste nerobili zlé meno našej fakulte. | ||
| + | * V projekte môžete porovnávať niekoľko programov na tú istú úlohu z hľadiska ich rýchlosti či presnosti výsledkov. Obsahom projektu bude príprava dát, na ktorých budete programy bežať, samotné spúšťanie (vhodne zoskriptované) ako aj vyhodnotenie výsledkov. | ||
| + | * A samozrejme môžete niekde na internete vyhrabať zaujímavé dáta a snažiť sa z nich niečo vydolovať. | ||
| + | |||
| + | ==Opisovanie== | ||
| + | |||
| + | * Máte povolené sa so spolužiakmi a ďalšími osobami rozprávať o domácich úlohách resp. projektoch a stratégiách na ich riešenie. Kód, získané výsledky aj text, ktorý odovzdáte, musí však byť vaša samostatná práca. Je zakázané ukazovať svoj kód alebo texty spolužiakom. | ||
| + | |||
| + | * Pri riešení domácej úlohy a projektu očakávame, že budete využívať internetové zdroje, najmä rôzne manuály a diskusné fóra k preberaným technológiám. Nesnažte sa však nájsť hotové riešenia zadaných úloh. Všetky použité zdroje uveďte v domácich úlohách a projektoch. | ||
| + | |||
| + | * Ak nájdeme prípady opisovania alebo nepovolených pomôcok, všetci zúčastnení študenti získajú za príslušnú domácu úlohu, projekt a pod. nula bodov (t.j. aj tí, ktorí dali spolužiakom odpísať) a prípad ďalej podstúpime na riešenie disciplinárnej komisii fakulty. | ||
| + | |||
| + | ==Zverejňovanie== | ||
| + | |||
| + | Zadania a materiály k predmetu sú voľne prístupné na tejto stránke. Prosím vás ale, aby ste nezverejňovali ani inak nešírili vaše riešenia domácich úloh, ak nie je v zadaní povedané inak. Vaše projekty môžete zverejniť, pokiaľ to nie je v rozpore s vašou dohodou so zadávateľom projektu a poskytovateľom dát. | ||
| + | =L01= | ||
| + | =Lecture 1: Perl, part 1= | ||
| + | |||
| + | ==Why Perl== | ||
| + | * From [https://en.wikipedia.org/wiki/Perl Wikipedia:] It has been nicknamed "the Swiss Army chainsaw of scripting languages" because of its flexibility and power, and possibly also because of its "ugliness". | ||
| + | |||
| + | Oficial slogans: | ||
| + | * There's more than one way to do it | ||
| + | * Easy things should be easy and hard things should be possible | ||
| + | |||
| + | Advantages | ||
| + | * Good capabilities for processing text files, regular expressions, running external programs etc. | ||
| + | * Closer to common programming language than shell scripts | ||
| + | * Perl one-liners on the command line can replace many other tools such as sed and awk | ||
| + | * Many existing libraries | ||
| + | |||
| + | Disadvantages | ||
| + | * Quirky syntax | ||
| + | * It is easy to write very unreadable programs (sometimes joking called write-only language) | ||
| + | * Quite slow and uses a lot of memory. If possible do no read entire input to memory, process line by line | ||
| + | |||
| + | Warning: we will use Perl 5, Perl 6 is quite a different language | ||
| + | |||
| + | ==Sources of Perl-related information== | ||
| + | * In package perl-doc man pages: | ||
| + | ** '''man perlintro''' introduction to Perl | ||
| + | ** '''man perlfunc''' list of standard functions in Perle | ||
| + | ** '''perldoc -f split''' describes function split, similarly other functions | ||
| + | ** '''perldoc -q sort''' shows answers to commonly asked questions (FAQ) | ||
| + | ** '''man perlretut''' and '''man perlre''' regular expressions | ||
| + | ** '''man perl''' list of other manual pages about Perl | ||
| + | * The same content on the web http://perldoc.perl.org/ | ||
| + | * Various web tutorials e.g. [http://www.perl.com/pub/a/2000/10/begperl1.html this one] | ||
| + | * Books | ||
| + | ** Simon Cozens: Beginning Perl [http://www.perl.org/books/beginning-perl/] freely downloadable | ||
| + | ** Larry Wall et al: Programming Perl [http://oreilly.com/catalog/9780596000271/] classics, Camel book | ||
| + | * '''Bioperl''' [http://www.bioperl.org/wiki/Main_Page] big library for bioinformatics | ||
| + | * Perl for Windows: http://strawberryperl.com/ | ||
| + | |||
| + | ==Hello world== | ||
| + | It is possible to run the code directly from a command line (more later): | ||
| + | <pre> | ||
| + | perl -e'print "Hello world\n"' | ||
| + | </pre> | ||
| + | |||
| + | This is equivalen to the following code stored in a file: | ||
| + | <pre> | ||
| + | #! /usr/bin/perl -w | ||
| + | use strict; | ||
| + | print "Hello world!\n"; | ||
| + | </pre> | ||
| + | |||
| + | * First line is a path to the interpreter | ||
| + | * Swith -w switches warnings on, e.g. if we manipulate with an undefined value (equivalen to "use warnings;") | ||
| + | * Second line <tt>use strict</tt> will switch on a more strict syntax checks, e.g. all variables must be defined | ||
| + | * Use of -w and use strict is strongly recommended | ||
| + | |||
| + | * Store the program in a file, e.g. <tt>hello.pl</tt> | ||
| + | * Make it executable (<tt>chmod a+x hello.pl</tt>) | ||
| + | * Run it with command <tt>./hello.pl</tt> | ||
| + | * Also possible to run as <tt>perl hello.pl</tt> (e.g. if we don't have the path to the interpreter in the file or the executable bit set) | ||
| + | |||
| + | ==The first input file for today: sequence repeats== | ||
| + | * In genomes some sequences occur in many copies (often not exactly equal, only similar) | ||
| + | * We have downloaded a table containing such sequence repeats on chromosome 2L of the fruitfly Drosophila melanogaster | ||
| + | * It was done as follows: on webpage http://genome.ucsc.edu/ we select drosophila genome, then in main menu select Tools, Table browser, select group: variation and repeats, track: ReapatMasker, region: position chr2L, output format: all fields from the selected table a output file: repeats.txt | ||
| + | * Each line of the file contains data about one repeat in the selected chromosome. The first line contains column names. Columns are tab-separated. Here are the first two lines: | ||
| + | <pre> | ||
| + | #bin swScore milliDiv milliDel milliIns genoName genoStart genoEnd genoLeft strand repName repClass repFamily repStart repEnd repLeft id | ||
| + | 585 778 167 7 20 chr2L 1 154 -23513558 + HETRP_DM Satellite Satellite 1519 1669 -203 1 | ||
| + | </pre> | ||
| + | * The file can be found at our server under filename <tt>/tasks/hw01/repeats.txt</tt> (17185 lines) | ||
| + | * A small randomly selected subset of the table rows is in file <tt>/tasks/hw01/repeats-small.txt</tt> (159 lines) | ||
| + | |||
| + | ==A sample Perl program== | ||
| + | For each type of repeat (column 11 of the file when counting from 0) we want to compute the number of repeats of this type | ||
| + | <pre> | ||
| + | #!/usr/bin/perl -w | ||
| + | use strict; | ||
| + | |||
| + | #associative array (hash), with repeat type as key | ||
| + | my %count; | ||
| + | |||
| + | while(my $line = <STDIN>) { # read every line on input | ||
| + | chomp $line; # delete end of line, if any | ||
| + | |||
| + | if($line =~ /^#/) { # skip commented lines | ||
| + | next; # similar to "continue" in C, move to next iteration | ||
| + | } | ||
| + | |||
| + | # split the input line to columns on every tab, store them in an array | ||
| + | my @columns = split "\t", $line; | ||
| + | |||
| + | # check input - should have at least 17 columns | ||
| + | die "Bad input '$line'" unless @columns >= 17; | ||
| + | |||
| + | my $type = $columns[11]; | ||
| + | |||
| + | # increase counter for this type | ||
| + | $count{$type}++; | ||
| + | } | ||
| + | |||
| + | # write out results, types sorted alphabetically | ||
| + | foreach my $type (sort keys %count) { | ||
| + | print $type, " ", $count{$type}, "\n"; | ||
| + | } | ||
| + | </pre> | ||
| + | |||
| + | This program does the same thing as the following one-liner (more on one-liners in two weeks) | ||
| + | <pre> | ||
| + | perl -F'"\t"' -lane 'next if /^#/; die unless @F>=17; $count{$F[11]}++; END { foreach (sort keys %count) { print "$_ $count{$_}" }}' filename | ||
| + | </pre> | ||
| + | |||
| + | ==Variables, types== | ||
| + | |||
| + | ===Scalar variables=== | ||
| + | * Scalar variables start with $, they can hold undefined value (<tt>undef</tt>), string, number, reference etc. | ||
| + | * Perl converts automatically between strings and numbers | ||
| + | <pre> | ||
| + | perl -e'print((1 . "2")+1, "\n")' | ||
| + | 13 | ||
| + | perl -e'print(("a" . "2")+1, "\n")' | ||
| + | 1 | ||
| + | perl -we'print(("a" . "2")+1, "\n")' | ||
| + | Argument "a2" isn't numeric in addition (+) at -e line 1. | ||
| + | 1 | ||
| + | </pre> | ||
| + | * If we switch on strict parsing, each variable needs to be defined by my, several variables created and initialized as follows: <tt>my ($a,$b) = (0,1);</tt> | ||
| + | * Usual set of C-style [http://perldoc.perl.org/perlop.html operators], power is **, string concatenation . | ||
| + | * Numbers compared by <, <=, ==, != etc., strings by lt, le, eq, ne, gt, ge, | ||
| + | * Comparison operator $a cmp $b for strings, $a <=> $b for numbers: returns -1 if $a<$b, 0 if they are equal, +1 if $a>$b | ||
| + | |||
| + | ===Arrays=== | ||
| + | * Names start with @, e.g. @a | ||
| + | * Access to element 0 in array: $a[0] | ||
| + | ** Starts with $, because the expression as a whole is a scalar value | ||
| + | * Length of array <tt>scalar(@a)</tt>. In scalar context, @a is the same thing. | ||
| + | ** e.g. <tt>for(my $i=0; $i<@a; $i++) { ... }</tt> | ||
| + | * If using non-existent indexes, they will be created, initialized to undef (++, += treat undef as 0) | ||
| + | * Stack/vector using functions push and pop: push @a, (1,2,3); $x = pop @a; | ||
| + | * Analogicaly shift and unshift on the left end of the array (slower) | ||
| + | * Sorting | ||
| + | ** @a = sort @a; (sorts alphabetically) | ||
| + | ** @a = sort {$a <=> $b} @a; (sort numerically) | ||
| + | ** { } can contain arbitrary comparison function, $a and $b are the two compared elements | ||
| + | * Array concatenation @c = (@a,@b); | ||
| + | * Swap values of two variables: ($x,$y) = ($y,$x); | ||
| + | * Iterate through values of an array (values can be changed): | ||
| + | <pre> | ||
| + | perl -e'my @a = (1,2,3); foreach my $val (@a) { $val++; } print join(" ", @a), "\n";' | ||
| + | 2 3 4 | ||
| + | </pre> | ||
| + | |||
| + | ===Associative array (hashes)=== | ||
| + | * Names start with %, e.g. %b | ||
| + | * Access elemtn with name "X": $b{"X"} | ||
| + | * Write out all elements of associative array %b | ||
| + | <pre> | ||
| + | foreach my $key (keys %b) { | ||
| + | print $key, " ", $b{$key}, "\n"; | ||
| + | } | ||
| + | </pre> | ||
| + | * Inicialization with constant: %b = ("key1"=>"value1","key2"=>"value2") | ||
| + | ** instead of => you can also use , | ||
| + | * test for existence of a key: if(exists $a{"x"}) {...} | ||
| + | * (other methods will create the queried key with undef value) | ||
| + | |||
| + | ===Multidimensional arrays, fun with pointers=== | ||
| + | * Pointer to a variable: \$a, \@a, \%a | ||
| + | * Pointer to an anonymous array: [1,2,3], pointer to an anonymous hash: {"kluc1"=>"hodnota1"} | ||
| + | * Hash of lists: | ||
| + | <pre> | ||
| + | my %a = ("fruits"=>["apple","banana","orange"], "vegetables"=>["celery","carrot"]} | ||
| + | $x = $a{"fruits"}[1]; | ||
| + | push @{$a{"fruits"}}, "kiwi"; | ||
| + | my $aref = \%a; | ||
| + | $x = $aref->{"fruits"}[1]; | ||
| + | </pre> | ||
| + | * Module Data::Dumper has function Dumper, which will recursively print complex data structures | ||
| + | |||
| + | ==Strings, regular expressions== | ||
| + | ===Strings=== | ||
| + | * Substring: <tt>[http://perldoc.perl.org/functions/substr.html substr]($string, $start, $length)</tt> | ||
| + | ** used also to access individual charaters (use length 1) | ||
| + | ** If we omit $length, considers until the end of the string, negative start counted from the end of the stringzaciatok rata od konca,... | ||
| + | ** We can also used to replace a substring by something else: <tt>substr($str, 0, 1) = "aaa"</tt> (replaces the first character by "aaa") | ||
| + | * Length of a string: <tt>[http://perldoc.perl.org/functions/length.html length]($str)</tt> | ||
| + | * Splitting a string to parts: <tt>[http://perldoc.perl.org/functions/split.html split] reg_expression, $string, $max_number_of_parts</tt> | ||
| + | ** if " " instead of regular expression, splits at whitespace | ||
| + | * Connecting parts <tt>[http://perldoc.perl.org/functions/join.html join]($separator, @strings)</tt> | ||
| + | * Other useful functions: <tt>[http://perldoc.perl.org/functions/chomp.html chomp]</tt> (removes end of line), <tt>[http://perldoc.perl.org/functions/index.html index]</tt> (finds a substring), lc, uc (conversion to lowercase/uppercase), reverse (mirror image), sprintf (C-style formatting) | ||
| + | |||
| + | ===Regular expressions=== | ||
| + | * more in [http://perldoc.perl.org/perlretut.html] | ||
| + | <pre> | ||
| + | $line =~ s/\s+$//; # remove whitespace at the end of the line | ||
| + | $line =~ s/[0-9]+/X/g; # replace each sequence of numbers with character X | ||
| + | |||
| + | #from the name of the fasta sequence (starting with >) create a string until the first space | ||
| + | #(\S means non-whitespace), the result is stored in $1, as specified by () | ||
| + | if($line =~ /^\>(\S+)/) { $name = $1; } | ||
| + | |||
| + | perl -le'$X="123 4 567"; $X=~s/[0-9]+/X/g; print $X' | ||
| + | X X X | ||
| + | </pre> | ||
| + | |||
| + | ==Conditionals, loops== | ||
| + | <pre> | ||
| + | if(expression) { # [] and () cannot be omitted | ||
| + | commands | ||
| + | } elsif(expression) { | ||
| + | commands | ||
| + | } else { | ||
| + | commands | ||
| + | } | ||
| + | |||
| + | command if expression; # here () not necessary | ||
| + | command unless expression; | ||
| + | die "negative value of x: $x" unless $x>=0; | ||
| + | |||
| + | for(my $i=0; $i<100; $i++) { | ||
| + | print $i, "\n"; | ||
| + | } | ||
| + | |||
| + | foreach my $i (0..99) { | ||
| + | print $i, "\n"; | ||
| + | } | ||
| + | |||
| + | $x=1; | ||
| + | while(1) { | ||
| + | $x *= 2; | ||
| + | last if $x>=100; | ||
| + | } | ||
| + | </pre> | ||
| + | |||
| + | * Undefined value, number 0 and strings "" and "0" evaluate as false, but I would recommmend always explicitly using logical values in conditional expressions, e.g. if(defined $x), if($x eq ""), if($x==0) etc. | ||
| + | |||
| + | ==Input, output== | ||
| + | * Reading one line from standard input: <tt>$line = <STDIN></tt> | ||
| + | * If no more input data available, returns undef | ||
| + | * See also [http://perldoc.perl.org/perlop.html#I%2fO-Operators] | ||
| + | * Special idiom <tt>while(my $line = <STDIN>)</tt> equivalent to <tt>while (defined(my $line = <STDIN>))</tt> | ||
| + | ** iterates through all lines of input | ||
| + | * <tt>chomp $line</tt> removes "\n", if any from the end of the string | ||
| + | * output to stdout through <tt>[http://perldoc.perl.org/functions/print.html print]</tt> or <tt>[http://perldoc.perl.org/functions/printf.html printf]</tt> | ||
| + | |||
| + | ==The second input file for today: DNA sequencing reads (fastq)== | ||
| + | |||
| + | * DNA sequencing machines can read only short pieces of DNA called reads | ||
| + | * Reads are usually stored in [https://en.wikipedia.org/wiki/FASTQ_format fastq format] | ||
| + | * Files can be very large (gigabytes or more), but we will use only a small sample from bacteria Staphylococcus aureus, source [http://gage.cbcb.umd.edu/data/Staphylococcus_aureus/Data.original/] | ||
| + | * Each read is on 4 lines: | ||
| + | ** line 1: ID of the read and other description, line starts with @ | ||
| + | ** line 2: DNA sequence, A,C,G,T are bases (nucleotides) of DNA, N means unknown base | ||
| + | ** line 3: + | ||
| + | ** line 4: quality string, which is the string of the same length as DNA in line 2. Each character represents quality of one base in DNA. If p is the probability that this base is wrong, the quality string will contain character with ASCII value 33+(-10 log p), where log is decimal logarithm. This means that higher ASCII means base of higher quality. Character ! (ASCII 33) means probability 1 of error, character $ (ASCII 36) means 50% error, character + (ASCII 43) is 10% error, character 5 (ASCII 53) is 1% error. | ||
| + | ** Note that some sequencing platforms represent qualities differently (see article linked above) | ||
| + | * Our file has all reads of equal length (this is not always the case) | ||
| + | |||
| + | The first 4 reads from file /tasks/hw01/reads-small.fastq | ||
| + | <pre> | ||
| + | @SRR022868.1845/1 | ||
| + | AAATTTAGGAAAAGATGATTTAGCAACATTTAGCCTTAATGAAAGACCAGATTCTGTTGCCATGTTTGAATGCCTTAAACCAGTAGCAGAATCAGTATAAA | ||
| + | + | ||
| + | IICIIIIIIIIIID%IIII8>I8III1II,II)I+III*II<II,E;-HI>+I0IB99I%%2GI*=?5*&1>'$0;%'+%%+;#'$&'%%$-+*$--*+(% | ||
| + | @SRR022868.1846/1 | ||
| + | TAGCGTTGTAAAATAAATTTCTAGAATGGAAGTGATGATATTGAAATACACTCAGATCCTGAATGAAAGATTTATTAAAGTTAAGACGAGAGTCTCATTAT | ||
| + | + | ||
| + | 4CIIIIIIII52I)IIIII0I16IIIII2IIII;IIAII&I6AI+*+&G5&G.@8/6&%&,03:*.$479.91(9--$,*&/3"$#&*'+#&##&$(&+&+ | ||
| + | </pre> | ||
| + | |||
| + | And now start on [[#HW01]] | ||
| + | =HW01= | ||
| + | See [[#L01|Lecture 1]] | ||
| + | |||
| + | ==Files== | ||
| + | We have 4 input files for this homework. We recommend creating soft links to your working directory as follows: | ||
| + | <pre> | ||
| + | ln -s /tasks/hw01/repeats-small.txt . # small version of the repeat file | ||
| + | ln -s /tasks/hw01/repeats.txt . # full version of the repeat file | ||
| + | ln -s /tasks/hw01/reads-small.fastq . # smaller version of the read file | ||
| + | ln -s /tasks/hw01/reads.fastq . # bigger version of the read file | ||
| + | </pre> | ||
| + | |||
| + | We recommend writing your protocol starting from an outline provided in <tt>/tasks/hw01/protocol.txt</tt> | ||
| + | |||
| + | ==Submitting== | ||
| + | * Directory /submit/hw01/your_username will be created for you | ||
| + | * Copy required files to this directory, including the protocol named protocol.txt or protocol.pdf | ||
| + | * You can modify these files freely until deadline, but after the deadline of the homework, you will lose access rights to this directory | ||
| + | |||
| + | ==Task A== | ||
| + | |||
| + | * Consider the program for counting repeat types in the [[#L01|lecture 1]], save it to file <tt>repeat-stat.pl</tt> | ||
| + | * Extend it to compute the average length of each type of repeat | ||
| + | ** Each row of the input table contains the start and end coordinates of the repeat in columns 7 and 6. The length is simply the difference of these two values. | ||
| + | * Output a table with three columns: type of repeat, the number of occurrences, the average length of the repeat. | ||
| + | ** Use printf to print these three items right-justified in columns of sufficient width, print the average length to 1 decimal place. | ||
| + | * If you run your script on the small file, the output should look something like this (exact column widths may differ): | ||
| + | <pre> | ||
| + | ./repeat-stat.pl < repeats-small.txt | ||
| + | DNA 5 377.4 | ||
| + | LINE 4 410.2 | ||
| + | LTR 13 355.4 | ||
| + | Low_complexity 22 47.2 | ||
| + | RC 8 236.2 | ||
| + | Simple_repeat 106 39.0 | ||
| + | </pre> | ||
| + | * Include in your '''protocol''' the output when you run your script on the large file: <tt>./repeat-stat.pl < repeats.txt</tt> | ||
| + | * Find out on [https://en.wikipedia.org/wiki/Retrotransposon Wikipedia], what acronyms LINE and LTR stand for. Do their names correspond to their lengths? (Write a short answer in the '''protocol'''.) | ||
| + | * '''Submit''' only your script, <tt>repeat-stat.pl</tt> | ||
| + | |||
| + | ==Task B== | ||
| + | |||
| + | * Write a script which reformats FASTQ file to FASTA format, call it <tt>fastq2fasta.pl</tt> | ||
| + | ** fastq file should be on standard input, fasta file written to standard output | ||
| + | * [https://en.wikipedia.org/wiki/FASTA_format FASTA format] is a typical format for storing DNA and protein sequences. | ||
| + | ** Each sequence consists of several lines of the file. The first line starts with ">" followed by identifier of the sequence and optinally some further description separated by whitespace | ||
| + | ** The sequence itself is on the second line, long sequences are split into multiple lines | ||
| + | * In our case, the name of the sequence will be the ID of the read with @ replaced by > and / replaced by _ | ||
| + | ** you can try to use [http://perldoc.perl.org/perlop.html#Quote-Like-Operators tr or s operators] | ||
| + | * For example, the first two reads of reads.fastq are: | ||
| + | <pre> | ||
| + | @SRR022868.1845/1 | ||
| + | AAATTTAGGAAAAGATGATTTAGCAACATTTAGCCTTAATGAAAGACCAGATTCTGTTGCCATGTTTGAATGCCTTAAACCAGTAGCAGAATCAGTATAAA | ||
| + | + | ||
| + | IICIIIIIIIIIID%IIII8>I8III1II,II)I+III*II<II,E;-HI>+I0IB99I%%2GI*=?5*&1>'$0;%'+%%+;#'$&'%%$-+*$--*+(% | ||
| + | @SRR022868.1846/1 | ||
| + | TAGCGTTGTAAAATAAATTTCTAGAATGGAAGTGATGATATTGAAATACACTCAGATCCTGAATGAAAGATTTATTAAAGTTAAGACGAGAGTCTCATTAT | ||
| + | + | ||
| + | 4CIIIIIIII52I)IIIII0I16IIIII2IIII;IIAII&I6AI+*+&G5&G.@8/6&%&,03:*.$479.91(9--$,*&/3"$#&*'+#&##&$(&+&+ | ||
| + | </pre> | ||
| + | * These should be reformatted as follows: | ||
| + | <pre> | ||
| + | >SRR022868.1845_1 | ||
| + | AAATTTAGGAAAAGATGATTTAGCAACATTTAGCCTTAATGAAAGACCAGATTCTGTTGCCATGTTTGAATGCCTTAAACCAGTAGCAGAATCAGTATAAA | ||
| + | >SRR022868.1846_1 | ||
| + | TAGCGTTGTAAAATAAATTTCTAGAATGGAAGTGATGATATTGAAATACACTCAGATCCTGAATGAAAGATTTATTAAAGTTAAGACGAGAGTCTCATTAT | ||
| + | </pre> | ||
| + | * '''Submit''' files <tt>fastq2fasta.pl</tt> and <tt>reads-small.fasta</tt> | ||
| + | ** the latter file is created by running <tt>./fastq2fasta.pl < reads-small.fastq > reads-small.fasta</tt> | ||
| + | |||
| + | ==Task C== | ||
| + | |||
| + | * Write a script <tt>fastq-quality.pl</tt> which for each position in a read computes the average quality | ||
| + | * Standard input has fastq file with multiple reads, possibly of different lengths | ||
| + | * As quality we will use simply ASCII values of characters in the quality string with value 33 substracted, so the quality is -10 log p | ||
| + | ** ASCII value can be computed by function [http://perldoc.perl.org/functions/ord.html ord] | ||
| + | * Positions in reads will be numbered from 0 | ||
| + | * Since reads can differ in length, some positions are used in more reads, some in fewer | ||
| + | * For each position from 0 up to the highest position used in some read, print three numbers separated by tabs "\t": the position index, the number of times this position was used in reads, average quality at that position with 1 decimal place (you can again use printf) | ||
| + | * The last two lines when you run <tt>./fastq-quality.pl < reads-small.fastq</tt> should be | ||
| + | <pre> | ||
| + | 99 86 5.5 | ||
| + | 100 86 8.6 | ||
| + | </pre> | ||
| + | * Run the following command, which runs your script on the larger file and selects every 10th position. Include the output in your '''protocol'''. Do you see any trend in quality values with increasing position? (Include a short comment in '''protocol'''.) | ||
| + | <pre> | ||
| + | ./fastq-quality.pl < reads.fastq | perl -lane 'print if $F[0]%10==0' | ||
| + | </pre> | ||
| + | * '''Submit''' only <tt>fastq-quality.pl</tt> | ||
| + | |||
| + | ==Task D== | ||
| + | |||
| + | * Write script <tt>fastq-trim.pl</tt> that trims low quality bases from the end of each read and filters out short reads | ||
| + | * This script should read a fastq file from standard input and write trimmed fastq file to standard output | ||
| + | * It should also accept two command-line arguments: character Q and integer L | ||
| + | ** We have not covered processing command line arguments, but you can use the code snippet below for this | ||
| + | * Q is the minimum acceptable quality (characters from quality string with ASCII value >= ASCII value of Q are ok) | ||
| + | * L is the minimum acceptable length of a read | ||
| + | * First find the last base in a read which has quality at least Q (if any). All bases after this base will be removed from both sequence and quality string | ||
| + | * If the resulting read has fewer than L bases, it is omitted from the output | ||
| + | |||
| + | You can check your program by the following tests: | ||
| + | * If you run the following two commands, you should get tmp identical with input and thus output of diff should be empty | ||
| + | <pre> | ||
| + | ./fastq-trim.pl '!' 101 < reads-small.fastq > tmp # trim at quality ASCII >=33 and length >=101 | ||
| + | diff reads-small.fastq tmp # output should be empty (no differences) | ||
| + | </pre> | ||
| + | |||
| + | * If you run the following two commands, you should see differences in 4 reads, 2 bases trimmed from each | ||
| + | <pre> | ||
| + | ./fastq-trim.pl '"' 1 < reads-small.fastq > tmp # trim at quality ASCII >=34 and length >=1 | ||
| + | diff reads-small.fastq tmp # output should be differences in 4 reads | ||
| + | </pre> | ||
| + | |||
| + | * If you run the following commands, you should get empty output (no reads meet the criteria): | ||
| + | <pre> | ||
| + | ./fastq-trim.pl d 1 < reads-small.fastq # quality ASCII >=100, length >= 1 | ||
| + | ./fastq-trim.pl '!' 102 < reads-small.fastq # quality ASCII >=33 and length >=102 | ||
| + | </pre> | ||
| + | |||
| + | Further runs and submitting | ||
| + | * Run <tt>./fastq-trim.pl '(' 95 < reads-small.fastq > reads-small-filtered.fastq # quality ASCII >= 40</tt> | ||
| + | * '''Submit''' files <tt>fastq-trim.pl</tt> and <tt>reads-small-filtered.fastq</tt> | ||
| + | * If you have done task C, run quality statistics on a trimmed version of the bigger file using command below and include in the '''protocol''' the result. Comment in the '''protocol''' on differences between statistics on the whole file in part C and D. Are they as you expected? | ||
| + | <pre> | ||
| + | ./fastq-trim.pl 2 50 < reads.fastq | ./fastq-quality.pl | perl -lane 'print if $F[0]%10==0' # quality ASCII >= 50 | ||
| + | </pre> | ||
| + | * Note: you have created tools which can be combined, e.g. you can create quality-trimmed version of the fasta file by first trimming fastq and then converting to fasta (no need to submit these files) | ||
| + | |||
| + | Parsing command-line arguments in this task (they will be stored in variables $Q and $L): | ||
| + | <pre> | ||
| + | #!/usr/bin/perl -w | ||
| + | use strict; | ||
| + | |||
| + | my $USAGE = " | ||
| + | Usage: | ||
| + | $0 Q L < input.fastq > output.fastq | ||
| + | |||
| + | Trim from the end of each read bases with ASCII quality value less | ||
| + | than the given threshold Q. If the length of the read after trimming | ||
| + | is less than L, the read will be omitted from output. | ||
| + | |||
| + | L is a non-negative integer, Q is a character | ||
| + | "; | ||
| + | |||
| + | # check that we have exactly 2 command-line arguments | ||
| + | die $USAGE unless @ARGV==2; | ||
| + | # copy command-line arguments to variables Q and L | ||
| + | my ($Q, $L) = @ARGV; | ||
| + | # check that $Q is one character and $L looks like a non-negative integer | ||
| + | die $USAGE unless length($Q)==1 && $L=~/^[0-9]+$/; | ||
| + | </pre> | ||
| + | =L02= | ||
| + | ==Motivation: Building Phylogenetic Trees== | ||
| + | The task for today will be to build a [https://en.wikipedia.org/wiki/Phylogenetic_tree phylogenetic tree] of several species using sequences of several genes. | ||
| + | * A phylogenetic tree is a tree showing evolutionary history of these species. Leaves are target present-day species, internal nodes are their common ancestors. | ||
| + | * Input contains sequences of genes from each species. | ||
| + | * Step 1: Identify ''ortholog groups''. Orthologs are genes from different species that "correspond" to each other. This is done based on sequence similarity and we can use a tool called [http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=Download blast] to identify sequence similarities between individual genes. The result of ortholog group identification will be a set of genes, each gene having one sequence from each of the 6 species | ||
| + | chimp_94013 dog_84719 human_15749 macaque_34640 mouse_17461 rat_09232 | ||
| + | * Step 2: For each ortholog group, we need to align genes and build a phylogenetic tree for this gene using existing methods. We can do this using tools muscle (for alignment) and phyml (for phylogenetic tree inference). | ||
| + | |||
| + | Unaligned sequences: | ||
| + | >mouse | ||
| + | ATGCAGTTCCCGCACCCGGGGCCCGCGGCTGCGCCCGCCGTGGGAGTCCCGCTGTATGCG | ||
| + | >rat | ||
| + | ATGCAGTTCCCGCACCCGGGGCCCGCGGCTGCGCCCGCCGTCGGAGTCCCGCTGTACGCG | ||
| + | >dog | ||
| + | ATGCAGTACCACCCCGGGCCGGCGGCGGCGGGCGCCGTGGGGGTGCCGCTGTACGCG | ||
| + | >human | ||
| + | ATGCAGTACCCGCACCCCGGGCCGGCGGCGGGCGCCGTGGGGGTGCCGCTGTACGCG | ||
| + | >chimp | ||
| + | ATGCAGTACCCGCACCCCGGGCCGGCGGCGGGCGCCGTGGGGGTGCCGCTGTACGCG | ||
| + | >macaque | ||
| + | ATGCAGTACCCGCACCCCGGGCGGCGGCCGTGGGGGTGGC | ||
| + | |||
| + | Aligned sequences: | ||
| + | >mouse | ||
| + | ATGCAGTTCCCGCACCCGGGGCCCGCGGCTGCGCCCGCCGTGGGAGTCCCGCTGTATGCG | ||
| + | >rat | ||
| + | ATGCAGTTCCCGCACCCGGGGCCCGCGGCTGCGCCCGCCGTCGGAGTCCCGCTGTACGCG | ||
| + | >dog | ||
| + | ATGCAGTAC---CACCCCGGGCCGGCGGCGGCGGGCGCCGTGGGGGTGCCGCTGTACGCG | ||
| + | >human | ||
| + | ATGCAGTACCCGCACCCCGGGC---CGGCGGCGGGCGCCGTGGGGGTGCCGCTGTACGCG | ||
| + | >chimp | ||
| + | ATGCAGTACCCGCACCCCGGGC---CGGCGGCGGGCGCCGTGGGGGTGCCGCTGTACGCG | ||
| + | >macaque | ||
| + | ATGCAGTACCCGCACCCCGGGC----------GGCGGCCGTGGGGGTGGC---------- | ||
| + | |||
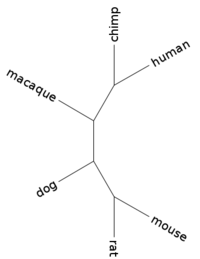
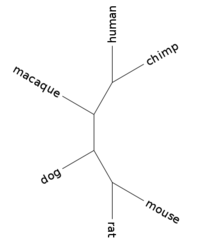
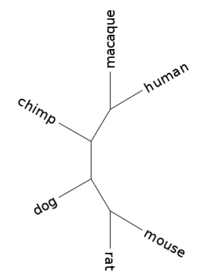
| + | Phylogenetic tree: | ||
| + | (mouse:0.03240286,rat:0.01544553,(dog:0.03632419,(macaque:0.01505050,(human:0.00000001,chimp:0.00000001):0.00627957):0.01396920):0.10645019); | ||
| + | |||
| + | [[Image:L02 human 15749.png|center|thumb|200px|Tree for gene human_15749 (branch lengths ignored)]] | ||
| + | |||
| + | |||
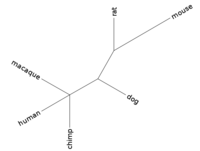
| + | * Step 3: The result of the previous step will be several trees, one for every gene. Ideally, all trees would be identical, showing the real evolutionary history of the six species. But it is not easy to infer the real tree from sequence data, so trees from different genes might differ. Therefore, in the last step, we will build a consensus tree. This can be done by usina interactive tool called phylip. | ||
| + | * Output is a single consensus tree. | ||
| + | |||
| + | <table> | ||
| + | <tr><td>[[Image:L02 human 15749.png|thumb|200px|Tree for gene human_15749]]</td> | ||
| + | <td>[[Image:L02 human 13531.png|thumb|200px|Tree for gene human_13531]]</td> | ||
| + | <td>[[Image:L02 human 31770.png|thumb|200px|Tree for gene human_31770]]</td></tr><tr> | ||
| + | <td>[[Image:L02 consensus.png|thumb|200px|Strict consensus for the three gene trees]]</td> | ||
| + | </tr></table> | ||
| + | |||
| + | |||
| + | Our goal for today is to build a pipeline that automates the whole task. | ||
| + | |||
| + | ==Opening files== | ||
| + | <pre> | ||
| + | my $in; | ||
| + | open $in, "<", "path/file.txt" or die; # open file for reading | ||
| + | while(my $line = <$in>) { | ||
| + | # process line | ||
| + | } | ||
| + | close $in; | ||
| + | |||
| + | my $out; | ||
| + | open $out, ">", "path/file2.txt" or die; # open file for writing | ||
| + | print $out "Hello world\n"; | ||
| + | close $out; | ||
| + | # if we want to append to a file use the following instead: | ||
| + | # open $out, ">>", "cesta/subor2.txt" or die; | ||
| + | |||
| + | # standard files | ||
| + | print STDERR "Hello world\n"; | ||
| + | my $line = <STDIN>; | ||
| + | # files as arguments of a function | ||
| + | citaj_subor($in); | ||
| + | citaj_subor(\*STDIN); | ||
| + | </pre> | ||
| + | |||
| + | ==Working with files and directories== | ||
| + | Working directories or files with automatically generated names are automagically deleted after the program finishes. | ||
| + | <pre> | ||
| + | use File::Temp qw/tempdir/; | ||
| + | my $dir = tempdir("atoms_XXXXXXX", TMPDIR => 1, CLEANUP => 1 ); | ||
| + | print STDERR "Creating temporary directory $dir\n"; | ||
| + | open $out,">$dir/myfile.txt" or die; | ||
| + | </pre> | ||
| + | |||
| + | Copying files | ||
| + | <pre> | ||
| + | use File::Copy; | ||
| + | copy("file1","file2") or die "Copy failed: $!"; | ||
| + | copy("Copy.pm",\*STDOUT); | ||
| + | move("/dev1/fileA","/dev2/fileB"); | ||
| + | </pre> | ||
| + | Other functions for working with file system, e.g. chdir, mkdir, unlink, chmod, ... | ||
| + | |||
| + | Function glob finds files with wildcard characters similarly as on command line (see also opendir, readdir, and File::Find module) | ||
| + | <pre> | ||
| + | ls *.pl | ||
| + | perl -le'foreach my $f (glob("*.pl")) { print $f; }' | ||
| + | </pre> | ||
| + | |||
| + | Additional functions for working with file names, paths, etc. in modules File::Spec and File::Basename. | ||
| + | |||
| + | Testing for an existence of a file (more in [http://perldoc.perl.org/functions/-X.html perldoc -f -X]) | ||
| + | <pre> | ||
| + | if(-r "file.txt") { ... } # is file.txt readable? | ||
| + | if(-d "dir") {.... } # is dir a directory? | ||
| + | </pre> | ||
| + | |||
| + | ==Running external programs== | ||
| + | <pre> | ||
| + | my $ret = system("command arguments"); | ||
| + | # returns -1 if cannot run command, otherwise pass the return code | ||
| + | </pre> | ||
| + | |||
| + | <pre> | ||
| + | my $allfiles = `ls`; | ||
| + | # returns the result of a command as a text | ||
| + | # cannot test return code | ||
| + | </pre> | ||
| + | |||
| + | Using pipes | ||
| + | <pre> | ||
| + | open $in, "ls |"; | ||
| + | while(my $line = <$in>) { ... } | ||
| + | </pre> | ||
| + | |||
| + | <pre> | ||
| + | open $out, "| wc"; | ||
| + | print $out "1234\n"; | ||
| + | close $out;' | ||
| + | |||
| + | 1 1 5 | ||
| + | </pre> | ||
| + | |||
| + | ==Command-line arguments== | ||
| + | <pre> | ||
| + | # module for processing options in a standardized way | ||
| + | use Getopt::Std; | ||
| + | # string with usage manual | ||
| + | my $USAGE = "$0 [options] length filename | ||
| + | |||
| + | Options: | ||
| + | -l switch on lucky mode | ||
| + | -o filename write output to filename | ||
| + | "; | ||
| + | |||
| + | # all arguments to the command are stored in @ARGV array | ||
| + | # parse options and remove them from @ARGV | ||
| + | my %options; | ||
| + | getopts("lo:", \%options); | ||
| + | # now there should be exactly two arguments in @ARGV | ||
| + | die $USAGE unless @ARGV==2; | ||
| + | # process options | ||
| + | my ($length, $filenamefile) = @ARGV; | ||
| + | # values of options are in the %options array | ||
| + | if(exists $options{'l'}) { print "Lucky mode\n"; } | ||
| + | </pre> | ||
| + | For long option names, see module Getopt::Long | ||
| + | |||
| + | ==Defining functions== | ||
| + | |||
| + | Defining new functions | ||
| + | <pre> | ||
| + | sub function_name { | ||
| + | # arguments are stored in @_ array | ||
| + | my ($firstarg, $secondarg) = @_; | ||
| + | # do something | ||
| + | return ($result, $second_result); | ||
| + | } | ||
| + | </pre> | ||
| + | * Arrays and hashes are usually passed as references: function_name(\@array, \%hash); | ||
| + | * It is advantageous to pass long string as references as well to prevent needless copying: function_name(\$sequence); | ||
| + | * References need to be dereferenced, e.g. substr($$sequence) or $array->[0] | ||
| + | |||
| + | ==Bioperl== | ||
| + | <pre> | ||
| + | use Bio::Tools::CodonTable; | ||
| + | sub translate | ||
| + | { | ||
| + | my ($seq, $code) = @_; | ||
| + | my $CodonTable = Bio::Tools::CodonTable->new( -id => $code); | ||
| + | my $result = $CodonTable->translate($seq); | ||
| + | |||
| + | return $result; | ||
| + | } | ||
| + | </pre> | ||
| + | |||
| + | |||
| + | ==Defining modules== | ||
| + | Module with name XXX should be in file XXX.pm. | ||
| + | <pre> | ||
| + | package shared; | ||
| + | |||
| + | BEGIN { | ||
| + | use Exporter (); | ||
| + | our (@ISA, @EXPORT, @EXPORT_OK); | ||
| + | @ISA = qw(Exporter); | ||
| + | # symbols to export by default | ||
| + | @EXPORT = qw(funkcia1, funkcia2); | ||
| + | } | ||
| + | |||
| + | sub funkcia1 { | ||
| + | ... | ||
| + | } | ||
| + | |||
| + | sub funkcia2 { | ||
| + | ... | ||
| + | } | ||
| + | |||
| + | #module must return true | ||
| + | 1; | ||
| + | </pre> | ||
| + | |||
| + | Using the module located in the same directory as .pl file: | ||
| + | <pre> | ||
| + | use FindBin qw($Bin); # $Bin is the directory with the script | ||
| + | use lib "$Bin"; # add bin to the library path | ||
| + | use shared; | ||
| + | </pre> | ||
| + | =HW02= | ||
| + | |||
| + | ==Biological background and overall approach== | ||
| + | The task for today will be to build a [https://en.wikipedia.org/wiki/Phylogenetic_tree phylogenetic tree] of several species using sequences of several genes. | ||
| + | * We will use 6 mammals: human, chimp, macaque, mouse, rat and dog | ||
| + | * A phylogenetic tree is a tree showing evolutionary history of these species. Leaves are target present-day species, internal nodes are their common ancestors. | ||
| + | * There are methods to build trees by comparing DNA or protein sequences of several present-day species. | ||
| + | * Our input contains a small selection of gene sequences from each species. In a real project we would start from all genes (cca 20,000 per species) and would do a careful filtration of problematic sequences, but we skip this step here. | ||
| + | * The first step will be to identify which genes from different species "correspond" to each other. More exactly, we are looking for groups of ''orthologs''. To do so, we will use a simple method based on sequence similarity, see details below. Again, in real project, more complex methods might be used. | ||
| + | * The result of ortholog group identification will be a set of genes, each gene having one sequence from each of the 6 species | ||
| + | * Next we will process each gene separately, aligning them and building a phylogenetic tree for this gene using existing methods. | ||
| + | * The result of the previous step will be several trees, one for every gene. Ideally, all trees would be identical, showing the real evolutionary history of the six species. But it is not easy to infer the real tree from sequence data, so trees from different genes might differ. Therefore, in the last step, we will build a consensus tree. | ||
| + | |||
| + | ==Technical overview== | ||
| + | |||
| + | This task can be organized in different ways, but to practice Perl, we will write a single Perl script which takes as an input a set of fasta files, each containing DNA sequences of several genes from a single species and writes on output the resulting consensus tree. | ||
| + | * For most of the steps, we will use existing bioinformatics tools. The script will run these tools and do some additional simple processing. | ||
| + | |||
| + | '''Temporary directory''' | ||
| + | * During its run, the script and various tools will generate many files. All these files will be stored in a single temporary directory which can be then easily deleted by the user. | ||
| + | * We will use Perl library [http://perldoc.perl.org/File/Temp.html File::Temp] to create this temporary directory with a unique name so that the script can be run several times simultaneously without clashing filenames. | ||
| + | * The library by default creates the file in /tmp, but instead we will create it in the current directory so that it is not deleted at restart of the computer and so that it can be more easily inspected for any problems | ||
| + | * The library by default deletes the directory when the script finishes but again, to allow inspection by the user, we will leave the directory in place | ||
| + | |||
| + | '''Restart''' | ||
| + | * The script will have a command line option for restarting the computation and omitting the time-consuming steps that were already finished | ||
| + | * This is useful in long-running scripts because during development of the script you will want to run it many times as you add more steps. In real usage the computation can also be interrupted for various reasons. | ||
| + | * Our restart capabilities will be quite rudimentary: before running a potentially slow external program, the script will check if the temporary directory contains a non-empty file with the filename matching the expected output of the program. If the file is found, it is assumed to be correct and complete and the external program is not run. | ||
| + | |||
| + | '''Command line options''' | ||
| + | * The script should be named build-tree.pl and as command-line arguments, it will get names of the species | ||
| + | ** For example, we can run the script as follows: <tt>./build-tree.pl human chimp macaque mouse rat dog</tt> | ||
| + | ** The first species, in this case human, will be so called reference species (see task A) | ||
| + | ** The script needs at least 2 species, otherwise it will write an error message and stop | ||
| + | ** For each species X there should be a file X.fa in the current directory, this is also checked by the script | ||
| + | * Restart is specified by command line option -r followed by the name of temporary directory | ||
| + | * Command-line option handling and creation of temporary directory is already implemented in the script you are given. | ||
| + | |||
| + | '''Input files''' | ||
| + | * Each input fasta X.fa file contains DNA sequences of several genes from one species X | ||
| + | * Each sequence name on a line starting with > will contain species name, underscore and gene id, e.g. ">human_00008" | ||
| + | * Species name matches name of the file, gene id is unique within the fasta file | ||
| + | * Species names and gene ids do not contain underscore, whitespace or any other special characters | ||
| + | * Sequence of each gene can be split into several lines | ||
| + | |||
| + | ==Files and submitting== | ||
| + | |||
| + | In /tasks/hw02/ you will find the following files: | ||
| + | * 6 fasta files (*.fa) | ||
| + | * skeleton script build-tree.pl | ||
| + | ** This script already contains handling of command line options, entire task B, potentially useful functions my_run and my_delete and suggested function headers for individual tasks. Feel free to change any of this. | ||
| + | * outline of protocol protocol.txt | ||
| + | * directory example with files for two different groups of genes | ||
| + | Copy the files to your directory and continue writing the script | ||
| + | |||
| + | Submitting | ||
| + | * Submit the script, protocol protocol.txt or protocol.pdf and temporary directory with all files created in the run of your script on all 6 species with human as reference. | ||
| + | * Since the commands and names of files are specified in the homework, you do not need to write them in the protocol (unless you change them). Therefore it is sufficient if the protocol contains self-assessment and any used information sources other than those linked from this assignment or lectures. | ||
| + | * Submit by copying to /submit/hw02/your_username | ||
| + | |||
| + | ==Task A: run blast to find similar sequences== | ||
| + | * To find orthologs, we use a simple method by first finding local alignments (regions of sequence similarity) between genes from different species | ||
| + | * For finding alignments, we will use tool [http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=Download blast] (ubuntu package blast2) | ||
| + | * Example of running blast: | ||
| + | |||
| + | formatdb -p F -i human.fa | ||
| + | blastall -p blastn -m 9 -d human.fa -i mouse.fa -e 1e-5 | ||
| + | |||
| + | * Example of output file: | ||
| + | |||
| + | # BLASTN 2.2.26 [Sep-21-2011] | ||
| + | # Query: mouse_00492 | ||
| + | # Database: human.fa | ||
| + | # Fields: Query id, Subject id, % identity, alignment length, mismatches, gap openings, q. start, q. end, s. start, s. end, e-value, bit score | ||
| + | mouse_22930 human_00008 90.79 1107 102 0 1 1107 1 1107 0.0 1386 | ||
| + | mouse_22930 human_34035 80.29 350 69 0 745 1094 706 1055 3e-37 147 | ||
| + | mouse_22930 human_34035 79.02 143 30 0 427 569 391 533 8e-07 46.1 | ||
| + | |||
| + | (note last column - score) | ||
| + | |||
| + | * For each non-reference ''species'', save the result of blast search in file ''species''.blast in the temporary directory. | ||
| + | |||
| + | ==Task B: find orthogroups== | ||
| + | '''This part is already implemented in the skeleton file, you don't need to implement or report anything in this task''' | ||
| + | * Here, we process all the '''species'''.blast files to find ortholog groups. | ||
| + | * Matches are symmetric, and there can be multiple matches for the same gene. We are looking for '''reciprocal best hits''': pairs of genes human_A and mouse_B, where mouse_B is the match with the highest score in mouse for human_A and human_A is the best-scoring match in human for mouse_B. | ||
| + | * Some genes in reference species may have no reciprocal best hits in some of the non-reference species. | ||
| + | * Gene in the reference species and all of its reciprocal best hits constitute '''orthogroup'''. If the size of an orthogroup is the same as the number of species, we will call it a '''complete orthogroup''' | ||
| + | * In file genes.txt in temporary directory list we will list all orthogroups, one per line. | ||
| + | chimp_94013 dog_84719 human_15749 macaque_34640 mouse_17461 rat_09232 | ||
| + | chimp_61053 human_18570 macaque_12627 | ||
| + | chimp_41364 human_19217 macaque_88256 rat_82436 | ||
| + | |||
| + | ==Task C: create a file for each orthogroup== | ||
| + | * For each complete orthogroup, we will create a fasta file with corresponding DNA sequences. | ||
| + | * The file will be located in temporary directory and will be named ''genename''.fa, where ''genename'' is the name of the orthogroup gene from reference species. | ||
| + | * The fasta name for each sequence is the name of species, NOT the name of the gene. | ||
| + | >human | ||
| + | CTGCGGCTGAGAGAGATGTGTACACTGGGGACGCACTCCGGATCTGCATAGTGACCAAAGAGGGCATCAGGGAGGAAACTGTTTCCTTAAGGAAGGAC | ||
| + | >chimp | ||
| + | TGCGGCTGAGAGAGATGTGTACACTGGGGACGCACTCCGGATCTGCATAGTGACCAAAGAGGGCATCAGGGAGGAGACTGTTTCCTTAAGGAAGGAC | ||
| + | >macaque | ||
| + | CTGCGGCTGAGAGAGACGTGTACACTGGGGACGCGCTCCGGATCTGCATAGTGACCAAAGAGGGCATCAGGGAGGAGACTGTTCCCTTAAGGAAGGAC | ||
| + | >mouse | ||
| + | CAGCCGAGAGGGATGTGTATACTGGAGATGCTCTCAGGATCTGCATCGTGACCAAAGAGGGCATCAGGGAGGAAACTGTTCCCCTGCGGAAAGAC | ||
| + | >rat | ||
| + | CAGCCGAGAGGGATGTGTACACTGGAGACGCCCTCAGGATCTGCATCGTGACCAAAGAGGGCATCAGGGAGGAGACTGTTCCCCTTCGGAAAGAC | ||
| + | >dog | ||
| + | GAGGGATGTGTACACTGGGGATGCACTCAGAATCTGCATTGTGACTAAGGAGGGCATCAGGGAGGAGACTGTTCCCCTGAGGAAGGAT | ||
| + | |||
| + | ==Task D: build tree for each gene== | ||
| + | * For each orthogroup, we need to build a phylogenetic tree. | ||
| + | * The result for file ''genename''.fa should be saved in file ''genename''.tree | ||
| + | * Example of how to do this: | ||
| + | # create multiple alignment of the sequences | ||
| + | muscle -diags -in genename.fa -out genename.mfa | ||
| + | # change format of the multiple alignment | ||
| + | readseq -f12 genename.mfa -o=genename.phy -a | ||
| + | # run phylogenetic inferrence program | ||
| + | phyml -i genename.phy --datatype nt --bootstrap 0 --no_memory_check | ||
| + | # rename the result | ||
| + | mv genename.phy_phyml_tree.txt genename.tree | ||
| + | * You can view the multiple alignment (*.mfa and *.phy) by using program seaview | ||
| + | * You can view the resulting tree (*.tree) by using program njplot or figtree | ||
| + | |||
| + | ==Task E: build consensus tree== | ||
| + | * Trees built on individual genes can differ from each other. | ||
| + | * Therefore we build a '''consensus tree''': tree that only contains branches present in most gene trees; other branches are collapsed. | ||
| + | * phylip is an "interactive" program for manipulation of trees. Specific command for [http://evolution.genetics.washington.edu/phylip/doc/consense.html building consensus trees] is | ||
| + | phylip consense | ||
| + | * input file for phylip needs to contain all trees of which consensus should be built, one per line | ||
| + | * text you would type to phylip manually, can be instead passed on the standard input from the script | ||
| + | * store the output tree from phylip in all_trees.consensus in temporary directory and also print it to standard output | ||
| + | =L03= | ||
| + | Today: using command-line tools and Perl one-liners. | ||
| + | * We will do simple transformations of text files using command-line tools without writing any scripts or longer programs. | ||
| + | * You will record the commands used in your protocol | ||
| + | ** We strongly recommend making a log of commands for data processing also outside of this course | ||
| + | * If you have a log of executed commands, you can easily execute them again by copy and paste | ||
| + | * For this reason any comments are best preceded by <tt>#</tt> | ||
| + | * If you use some sequence of commands often, you can turn it into a script | ||
| + | |||
| + | Most commands have man pages or are described within <tt>man bash</tt> | ||
| + | |||
| + | ==Efficient use of command line== | ||
| + | |||
| + | Some tips for bash shell: | ||
| + | * use ''tab'' key to complete command names, path names etc | ||
| + | ** tab completion can be customized [https://www.debian-administration.org/article/316/An_introduction_to_bash_completion_part_1] | ||
| + | * use ''up'' and ''down'' keys to walk through history of recently executed commands, then edit and resubmit chosen command | ||
| + | * press ''ctrl-r'' to search in the history of executed commands | ||
| + | * at the end of session, history stored in <tt>~/.bash_history</tt> | ||
| + | * command <tt>history -a</tt> appends history to this file right now | ||
| + | ** you can then look into the file and copy appropriate commands to your protocol | ||
| + | * various other history tricks, e.g. special variables [http://samrowe.com/wordpress/advancing-in-the-bash-shell/] | ||
| + | * <tt>cd -</tt> goes to previously visited directory, also see <tt>pushd</tt> and <tt>popd</tt> | ||
| + | * <tt>ls -lt | head</tt> shows 10 most recent files, useful for seeing what you have done last | ||
| + | |||
| + | Instead of bash, you can use more advanced command-line environments, e.g. [http://ipython.org/notebook.html iPhyton notebook] | ||
| + | |||
| + | ==Redirecting and pipes== | ||
| + | |||
| + | <pre> | ||
| + | # redirect standard output to file | ||
| + | command > file | ||
| + | |||
| + | # append to file | ||
| + | command >> file | ||
| + | |||
| + | # redirect standard error | ||
| + | command 2>file | ||
| + | |||
| + | # redirect file to standard input | ||
| + | command < file | ||
| + | |||
| + | # do not forget to quote > in other uses | ||
| + | grep '>' sequences.fasta | ||
| + | # (without quotes rewrites sequences.fasta) | ||
| + | |||
| + | # send stdout of command1 to stdin of command2 | ||
| + | command1 | command2 | ||
| + | |||
| + | # backtick operator executes command, | ||
| + | # removes trailing \n from stdout, substitutes to command line | ||
| + | # the following commands do the same thing: | ||
| + | head -n 2 file | ||
| + | head -n `echo 2` file | ||
| + | |||
| + | # redirect a string in ' ' to stdin of command head | ||
| + | head -n 2 <<< 'line 1 | ||
| + | line 2 | ||
| + | line 3' | ||
| + | |||
| + | # in some commands, file argument can be taken from stdin if denoted as - or stdin | ||
| + | # the following compares uncompressed version of file1 with file2 | ||
| + | zcat file1.gz | diff - file2 | ||
| + | </pre> | ||
| + | |||
| + | Make piped commands fail properly: | ||
| + | <pre> | ||
| + | set -o pipefail | ||
| + | </pre> | ||
| + | If set, the return value of a pipeline is the value of the last (rightmost) command to exit with a non-zero status, or zero if all commands in the pipeline exit successfully. This option is disabled by default, pipe then returns exit status of the rightmost command. | ||
| + | |||
| + | ==Text file manipulation== | ||
| + | ===Commands echo and cat (creating and printing files)=== | ||
| + | <pre> | ||
| + | # print text Hello and end of line to stdout | ||
| + | echo "Hello" | ||
| + | # interpret backslash combinations \n, \t etc: | ||
| + | echo -e "first line\nsecond\tline" | ||
| + | # concatenate several files to stdout | ||
| + | cat file1 file2 | ||
| + | </pre> | ||
| + | |||
| + | ===Commands head and tail (looking at start and end of files)=== | ||
| + | <pre> | ||
| + | # print 10 first lines of file (or stdin) | ||
| + | head file | ||
| + | some_command | head | ||
| + | # print the first 2 lines | ||
| + | head -n 2 file | ||
| + | # print the last 5 lines | ||
| + | tail -n 5 file | ||
| + | # print starting from line 100 (line numbering starts at 1) | ||
| + | tail -n +100 file | ||
| + | # print lines 81..100 | ||
| + | head -n 100 file | tail -n 20 | ||
| + | </pre> | ||
| + | * Docs: [http://www.gnu.org/software/coreutils/manual/html_node/head-invocation.html head], [http://www.gnu.org/software/coreutils/manual/html_node/tail-invocation.html tail] | ||
| + | |||
| + | ===Commands wc, ls -lh, od (exploring file stats and details)=== | ||
| + | <pre> | ||
| + | # prints three numbers: number of lines (-l), number of words (-w), number of bytes (-c) | ||
| + | wc file | ||
| + | |||
| + | # prints size of file in human-readable units (K,M,G,T) | ||
| + | ls -lh file | ||
| + | |||
| + | # od -a prints file or stdout with named characters | ||
| + | # allows checking whitespace and special characters | ||
| + | echo "hello world!" | od -a | ||
| + | # prints: | ||
| + | # 0000000 h e l l o sp w o r l d ! nl | ||
| + | # 0000015 | ||
| + | </pre> | ||
| + | * Docs: [http://www.gnu.org/software/coreutils/manual/html_node/wc-invocation.html wc], [http://www.gnu.org/software/coreutils/manual/html_node/ls-invocation.html ls], [http://www.gnu.org/software/coreutils/manual/html_node/od-invocation.html od] | ||
| + | |||
| + | ===Command grep (getting lines in files or stdin matching a regular expression)=== | ||
| + | <pre> | ||
| + | # -i ignores case (upper case and lowercase letters are the same) | ||
| + | grep -i chromosome file | ||
| + | # -c counts the number of matching lines in each file | ||
| + | grep -c '^[12][0-9]' file1 file2 | ||
| + | |||
| + | # other options (there is more, see the manual): | ||
| + | # -v print/count not matching lines (inVert) | ||
| + | # -n show also line numbers | ||
| + | # -B 2 -A 1 print 2 lines before each match and 1 line after match | ||
| + | # -E extended regular expressions (allows e.g. |) | ||
| + | # -F no regular expressions, set of fixed strings | ||
| + | # -f patterns in a file | ||
| + | # (good for selecting e.g. only lines matching one of "good" ids) | ||
| + | </pre> | ||
| + | * docs: [http://www.gnu.org/software/grep/manual/grep.html grep] | ||
| + | |||
| + | ===Commands sort, uniq=== | ||
| + | <pre> | ||
| + | # some useful options of sort: | ||
| + | # -g numeric sort | ||
| + | # -k which column(s) to use as key | ||
| + | # -r reverse (from largest values) | ||
| + | # -s stable | ||
| + | # -t fiels separator | ||
| + | |||
| + | # sorting first by column 2 numerically, in case of ties use column 1 | ||
| + | sort -k 1 file | sort -g -s -k 2,2 | ||
| + | |||
| + | # uniq outputs one line from each group of consecutive identical lines | ||
| + | # uniq -c adds the size of each group as the first column | ||
| + | # the following finds all unique lines and sorts them by frequency from most frequent | ||
| + | sort file | uniq -c | sort -gr | ||
| + | </pre> | ||
| + | * docs: [http://www.gnu.org/software/coreutils/manual/html_node/sort-invocation.html sort], [http://www.gnu.org/software/coreutils/manual/html_node/uniq-invocation.html uniq] | ||
| + | |||
| + | ===Commands diff, comm (comparing files)=== | ||
| + | |||
| + | [http://www.gnu.org/software/coreutils/manual/html_node/diff-invocation.html diff] compares two files, useful for manual checking | ||
| + | * useful options | ||
| + | ** -b (ignore whitespace differences) | ||
| + | ** -r for comparing directories | ||
| + | ** -q for fast checking for identity | ||
| + | ** -y show differences side-by-side | ||
| + | |||
| + | [http://www.gnu.org/software/coreutils/manual/html_node/comm-invocation.html comm] compares two sorted files | ||
| + | * writes 3 columns: | ||
| + | ** 1: lines occurring in the first file | ||
| + | ** 2: lines occurring in the second file | ||
| + | ** 3: lines occurring in both files | ||
| + | * some columns can be suppressed with -1, -2, -3 | ||
| + | * good for finding set intersections and differences | ||
| + | |||
| + | ===Commands cut, paste, join (working with columns)=== | ||
| + | * [http://www.gnu.org/software/coreutils/manual/html_node/cut-invocation.html cut] selects only some columns from file (perl/awk more flexible) | ||
| + | * [http://www.gnu.org/software/coreutils/manual/html_node/paste-invocation.html paste] puts 2 or more files side by side, separated by tabs or other character | ||
| + | * [http://www.gnu.org/software/coreutils/manual/html_node/join-invocation.html join] is a powerful tool for making joins and left-joins as in databases on specified columns in two files | ||
| + | |||
| + | ===Commands split, csplit (splitting files to parts)=== | ||
| + | * [http://www.gnu.org/software/coreutils/manual/html_node/split-invocation.html split] splits into fixed-size pieces (size in lines, bytes etc.) | ||
| + | * [http://www.gnu.org/software/coreutils/manual/html_node/csplit-invocation.html csplit] splits at occurrence of a pattern (e.g. fasta file into individual sequences) | ||
| + | <pre> | ||
| + | csplit sequences.fa '/^>/' '{*}' | ||
| + | </pre> | ||
| + | |||
| + | ==Programs sed and awk== | ||
| + | Both programs process text files line by line, allow to do various transformations | ||
| + | * awk newer, more advanced | ||
| + | * several examples below | ||
| + | * More info on wikipedia: [https://en.wikipedia.org/wiki/AWK awk], [https://en.wikipedia.org/wiki/Sed sed] | ||
| + | <pre> | ||
| + | # replace text "Chr1" by "Chromosome 1" | ||
| + | sed 's/Chr1/Chromosome 1/' | ||
| + | # prints first two lines, then quits (like head -n 2) | ||
| + | sed 2q | ||
| + | |||
| + | # print first and second column from a file | ||
| + | awk '{print $1, $2}' | ||
| + | |||
| + | # print the line if difference in first and second column > 10 | ||
| + | awk '{ if ($2-$1>10) print }' | ||
| + | |||
| + | # print lines matching pattern | ||
| + | awk '/pattern/ { print }' | ||
| + | |||
| + | # count lines | ||
| + | awk 'END { print NR }' | ||
| + | </pre> | ||
| + | |||
| + | ==Perl one-liners== | ||
| + | Instead of sed and awk, we will cover Perl one-liners | ||
| + | * more examples [http://www.math.harvard.edu/computing/perl/oneliners.txt], [https://blogs.oracle.com/ksplice/entry/the_top_10_tricks_of] | ||
| + | * documentation for Perl switches [http://perldoc.perl.org/perlrun.html] | ||
| + | <pre> | ||
| + | # -e executes commands | ||
| + | perl -e'print 2+3,"\n"' | ||
| + | perl -e'$x = 2+3; print $x, "\n"'; | ||
| + | |||
| + | # -n wraps commands in a loop reading lines from stdin or files listed as arguments | ||
| + | # the following is roughly the same as cat: | ||
| + | perl -ne'print' | ||
| + | # how to use: | ||
| + | perl -ne'print' < input > output | ||
| + | perl -ne'print' input1 input2 > output | ||
| + | # lines are stored in a special variable $_ | ||
| + | # this variable is default argument of many functions, | ||
| + | # including print, so print is the same as print $_ | ||
| + | |||
| + | # simple grep-like commands: | ||
| + | perl -ne 'print if /pattern/' | ||
| + | # simple regular expression modifications | ||
| + | perl -ne 's/Chr(\d+)/Chromosome $1/; print' | ||
| + | # // and s/// are applied by default to $_ | ||
| + | |||
| + | # -l removes end of line from each input line and adds "\n" after each print | ||
| + | # the following adds * at the end of each line | ||
| + | perl -lne'print $_, "*"' | ||
| + | |||
| + | # -a splits line into words separated by whitespace and stores them in array @F | ||
| + | # the next example prints difference in numbers stored in the second and first column | ||
| + | # (e.g. interval size if each line coordinates of one interval) | ||
| + | perl -lane'print $F[1]-$F[0]' | ||
| + | |||
| + | # -F allows to set separator used for splitting (regular expression) | ||
| + | # the next example splits at tabs | ||
| + | perl -F '"\t"' -lane'print $F[1]-$F[0]' | ||
| + | |||
| + | # -i replaces each file with a new transformed version (DANGEROUS!) | ||
| + | # the next example removes empty lines from all .txt files in the current directory | ||
| + | perl -lne 'print if length($_)>0' -i *.txt | ||
| + | # the following example replaces sequence of whitespace by exactly one space | ||
| + | # and removes leading and trailing spaces from lines in all .txt files | ||
| + | perl -lane 'print join(" ", @F)' -i *.txt | ||
| + | |||
| + | # END { commands } is run at the very end, after we finish reading input | ||
| + | # the following example computes the sum of interval lengths | ||
| + | perl -lane'$sum += $F[1]-$F[0]; END { print $sum; }' | ||
| + | # similarly BEGIN { command } before we start | ||
| + | |||
| + | # variable $. contains line number. $ARGV name of file or - for stdin | ||
| + | # the following prints filename and line number in front of every line | ||
| + | perl -ne'printf "%s.%d: %s", $ARGV, $., $_' file1 file2 | ||
| + | |||
| + | # moving files *.txt to have extension .tsv: | ||
| + | # first print commands | ||
| + | # then execute by hand or replace print with system | ||
| + | # mv -i asks if something is to be rewritten | ||
| + | ls *.txt | perl -lne '$s=$_; $s=~s/\.txt/.tsv/; print("mv -i $_ $s")' | ||
| + | ls *.txt | perl -lne '$s=$_; $s=~s/\.txt/.tsv/; system("mv -i $_ $s")' | ||
| + | </pre> | ||
| + | =HW03= | ||
| + | [[#L01|Lecture 1]], [[#L02|Lecture 2]], [[#L03|Lecture 3]] | ||
| + | |||
| + | * In this homework, use command-line tools or one-liners in Perl, awk or sed. Do not write any scripts or programs. | ||
| + | * Each task can be split into several stages and intermediate files written to disk, but you can also use pipelines to reduce the number of temporary files. | ||
| + | * Your commands should work also for other input files with the same format (do not try to generalize them too much, but also do not use very specific properties of a particular input, such as the number of lines etc.) | ||
| + | * Include all relevant used commands in your protocol and add a short description of your approach. | ||
| + | * Submit the protocol and required output files. | ||
| + | * Outline of the protocol is in <tt>/tasks/hw03/protocol.txt</tt>, submit to directory <tt>/submit/hw03/yourname</tt> | ||
| + | |||
| + | ==Bonus== | ||
| + | * If you are bored, you can try to write solution of Task B using as small number of characters as possible | ||
| + | * In the protocol, include both normal readable form and the condensed form | ||
| + | * Winner with the shortest set of commands gets some bonus points | ||
| + | |||
| + | ==Task A== | ||
| + | * <tt>/tasks/hw03/names.txt</tt> contains data about several people, one per line. | ||
| + | * Each line consists of given name(s), surname and email separated by spaces. | ||
| + | * Each person can have multiple given names (at least 1), but exactly one surname and one email. Email is always of the form <tt>username@uniba.sk</tt>. | ||
| + | * The task is to generate file <tt>passwords.csv</tt> which contains a randomly generated password for each of these users | ||
| + | * The output file has columns separated by commas ',' | ||
| + | * The first column contains username extracted from email address, second column surname, third column all given names and fourth column the randomly generated password | ||
| + | * '''Submit''' file <tt>passwords.csv</tt> with the result of your commands. | ||
| + | * In addition to commands that produce the desired file, also run some commands that could help you to detect potential problems in the input. | ||
| + | ** Examples of such problems include input lines with fewer than 3 columns, missing @ in email, or fields containing commas. | ||
| + | ** For every type of error, run a command which should have a different result for correct and incorrect files, and this output should be short even if the input file is long. In your protocol, explain what output you expect for correct and incorrect files (e.g. you can use grep to find incorrect lines and count their number - in a good file the number would be 0, in a bad file higher than 0) | ||
| + | * Such checks are not necessary in the other tasks, but in a real project it is a good idea to check your input and intermediate files. | ||
| + | |||
| + | Example line from input: | ||
| + | <pre> | ||
| + | Pavol Országh Hviezdoslav hviezdoslav32@uniba.sk | ||
| + | </pre> | ||
| + | |||
| + | Example line from output (password will differ): | ||
| + | <pre> | ||
| + | hviezdoslav32,Hviezdoslav,Pavol Országh,3T3Pu3un | ||
| + | </pre> | ||
| + | |||
| + | Hints: | ||
| + | * Passwords can be generated using <tt>pwgen</tt>, we also recommend using <tt>perl</tt>, <tt>wc</tt>, <tt>paste</tt>. | ||
| + | * In Perl, function [http://perldoc.perl.org/functions/pop.html pop] may be useful for manipulating @F and function [http://perldoc.perl.org/functions/join.html join] for connecting strings with a separator. | ||
| + | |||
| + | ==Task B== | ||
| + | |||
| + | '''File:''' | ||
| + | * <tt>/tasks/hw03/saccharomyces_cerevisiae.gff</tt> contains annotation of the yeast genome | ||
| + | ** Downloaded from http://yeastgenome.org/ on 2016-03-09, in particular from [http://downloads.yeastgenome.org/curation/chromosomal_feature/saccharomyces_cerevisiae.gff]. | ||
| + | ** It was further processed to omit DNA sequences from the end of file. | ||
| + | ** The size of the file is 5.6M. | ||
| + | * For easier work, link the file to your directory by <tt>ln -s /tasks/hw03/saccharomyces_cerevisiae.gff yeast.gff</tt> | ||
| + | * The file is in GFF3 format [http://www.sequenceontology.org/gff3.shtml] | ||
| + | * Lines starting with <tt>#</tt> are comments, other lines contain tab-separated data about one interval of some chromosome in the yeast genome | ||
| + | * Meaning of the first 5 columns: | ||
| + | ** column 0 chromosome name | ||
| + | ** column 1 source (can be ignored) | ||
| + | ** column 2 type of interval | ||
| + | ** column 3 start of interval (1-based coordinates) | ||
| + | ** column 4 end of interval (1-based coordinates) | ||
| + | * You can assume that these first 5 columns do not contain whitespace | ||
| + | |||
| + | '''Task:''' | ||
| + | * For each chromosome, the file contains a line which has in column 2 string <tt>chromosome</tt>, and the interval is the whole chromosome. | ||
| + | * To file <tt>chrosomes.txt</tt>, print a tab-separated list of chromosome names and sizes in the same order as in the input | ||
| + | * The last line of <tt>chromosomes.txt</tt> should list the total size of all chromosomes combined. | ||
| + | * '''Submit''' file <tt>chromosomes.txt</tt> | ||
| + | * Hint: tab is written in Perl as <tt>"\t"</tt>. Command <tt>cat</tt> may be useful. | ||
| + | * Your output should start and end as follows: | ||
| + | <pre> | ||
| + | chrI 230218 | ||
| + | chrII 813184 | ||
| + | ... | ||
| + | ... | ||
| + | chrXVI 948066 | ||
| + | chrmt 85779 | ||
| + | total 12157105 | ||
| + | </pre> | ||
| + | |||
| + | ==Task C== | ||
| + | * Continue processing file from task B. Print for each type of interval (column 2), how many times it occurs in the file. | ||
| + | * Sort from the most common to the least common interval types. | ||
| + | * Hint: commands <tt>sort</tt> and <tt>uniq</tt> will be useful. Do not forget to skip comments. | ||
| + | * '''Submit''' file <tt>types.txt</tt> with the output formatted as follows: | ||
| + | <pre> | ||
| + | 7058 CDS | ||
| + | 6600 mRNA | ||
| + | ... | ||
| + | ... | ||
| + | 1 telomerase_RNA_gene | ||
| + | 1 mating_type_region | ||
| + | 1 intein_encoding_region | ||
| + | |||
| + | </pre> | ||
| + | |||
| + | ==Task D== | ||
| + | '''Overall goal:''' | ||
| + | * Proteins from several well-studied yeast species were downloaded from database http://www.uniprot.org/ on 2016-03-09 | ||
| + | * We have also downloaded proteins from yeast Yarrowia lipolytica. We will pretend that nothing is known about these proteins (as if they were produced by gene finding program in a newly sequenced genome). | ||
| + | * We have run blast of known proteins vs. Y.lip. proteins. | ||
| + | * Now we want to find for each protein in Y.lip. its closest match among all known proteins. | ||
| + | |||
| + | '''Files:''' | ||
| + | * <tt>/tasks/hw03/known.fa</tt> is a fasta file with known proteins from several species | ||
| + | * <tt>/tasks/hw03/yarLip.fa</tt> is a fasta file with proteins from Y.lip. | ||
| + | * <tt>/tasks/hw03/known.blast</tt> is the result of running blast of <tt>yarLip.fa</tt> versus <tt>known.fa</tt> by these commands: | ||
| + | <pre> | ||
| + | formatdb -i known.fa | ||
| + | blastall -p blastp -d known.fa -i yarLip.fa -m 9 -e 1e-5 > known.blast | ||
| + | </pre> | ||
| + | * you can link these files to your directory as follows: | ||
| + | <pre> | ||
| + | ln -s /tasks/hw03/known.fa . | ||
| + | ln -s /tasks/hw03/yarLip.fa . | ||
| + | ln -s /tasks/hw03/known.blast . | ||
| + | </pre> | ||
| + | |||
| + | '''Step 1:''' | ||
| + | * Get the first (strongest) match for each query from <tt>known.blast</tt>. | ||
| + | * This can be done by printing the lines that are not comments but follow a comment line starting with #. | ||
| + | * In a perl one-liner, you can create a state variable which will remember if the previous line was a comment and based on that you decide of you print the current line. | ||
| + | * Instead of using perl, you can play with grep. Option -A 1 prints the matching lines as well as one line ofter each match | ||
| + | * Print only the first two columns separated by tab (name of query, name of target), sort the file by second column. | ||
| + | * '''Submit''' file best.tsv with the result | ||
| + | * File should start as follows: | ||
| + | <pre> | ||
| + | Q6CBS2 sp|B5BP46|YP52_SCHPO | ||
| + | Q6C8R4 sp|B5BP48|YP54_SCHPO | ||
| + | Q6CG80 sp|B5BP48|YP54_SCHPO | ||
| + | Q6CH56 sp|B5BP48|YP54_SCHPO | ||
| + | </pre> | ||
| + | |||
| + | '''Step 2:''' | ||
| + | * '''Submit''' file <tt>known.tsv</tt> which contains sequence names extracted from known.fa with leading <tt>></tt> removed | ||
| + | * This file should be sorted alphabetically. | ||
| + | * File should start as follows: | ||
| + | <pre> | ||
| + | sp|A0A023PXA5|YA19A_YEAST Putative uncharacterized protein YAL019W-A OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) GN=YAL019W-A PE=5 SV=1 | ||
| + | sp|A0A023PXB0|YA019_YEAST Putative uncharacterized protein YAR019W-A OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) GN=YAR019W-A PE=5 SV=1 | ||
| + | </pre> | ||
| + | |||
| + | '''Step 3:''' | ||
| + | * Use command [http://www.gnu.org/software/coreutils/manual/html_node/join-invocation.html join] to join the files <tt>best.tsv</tt> and <tt>known.tsv</tt> so that each line of <tt>best.tsv</tt> is extended with the text describing the corresponding target in <tt>known.tsv</tt> | ||
| + | * Use option <tt>-1 2</tt> to use second column of <tt>best.tsv</tt> as a key for joining | ||
| + | * The output of join may look as follows: | ||
| + | <pre> | ||
| + | sp|B5BP46|YP52_SCHPO Q6CBS2 Putative glutathione S-transferase C1183.02 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) GN=SPBC460.02c PE=3 SV=1 | ||
| + | sp|B5BP48|YP54_SCHPO Q6C8R4 Putative alpha-ketoglutarate-dependent sulfonate dioxygenase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) GN=SPBC460.04c PE=3 SV=1 | ||
| + | </pre> | ||
| + | * Further reformat the output so that query name goes first (e.g. <tt>Q6CBS2</tt>), followed by target name (e.g. <tt>sp|B5BP46|YP52_SCHPO</tt>), followed by the rest of the text, but remove all text after <tt>OS=</tt> | ||
| + | * Sort by query name | ||
| + | * '''Submit''' file <tt>best.txt</tt> with the result | ||
| + | * The output should start as follows: | ||
| + | <pre> | ||
| + | B5FVA8 tr|Q5A7D5|Q5A7D5_CANAL Lysophospholipase | ||
| + | B5FVB0 sp|O74810|UBC1_SCHPO Ubiquitin-conjugating enzyme E2 1 | ||
| + | B5FVB1 sp|O13877|RPAB5_SCHPO DNA-directed RNA polymerases I, II, and III subunit RPABC5 | ||
| + | </pre> | ||
| + | |||
| + | '''Note:''' | ||
| + | * not all Y.lip. are necessarily included in your final output (some proteins do not have blast match) | ||
| + | ** you can think how to find the list of such proteins, but this is not part of the assignment | ||
| + | * but files <tt>best.txt</tt> and <tt>best.tsv</tt> should have the same number of lines | ||
| + | =L04= | ||
| + | ==Job Scheduling== | ||
| + | |||
| + | * Some computing jobs take a lot of time: hours, days, weeks,... | ||
| + | * We do not want to keep a command-line window open the whole time; therefore we run such jobs in the background | ||
| + | * Simple commands to do it in Linux: batch, at, screen | ||
| + | * These commands run jobs immediately ([https://www.gnu.org/software/screen/manual/screen.html screen], [https://tmux.github.io/ tmux]) or at preset time (at), or when the computer becomes idle ([http://pubs.opengroup.org/onlinepubs/9699919799/utilities/batch.html batch]) | ||
| + | * Now we will concentrate on '''[https://en.wikipedia.org/wiki/Oracle_Grid_Engine Sun Grid Engine]''', a complex software for managing many jobs from many users on a cluster from multiple computers | ||
| + | * Basic workflow: | ||
| + | ** Submit a job (command) to a queue | ||
| + | ** The job waits in the queue until resources (memory, CPUs, etc.) become available on some computer | ||
| + | ** Then the job runs on the computer | ||
| + | ** Output of the job stored in files | ||
| + | ** User can monitor the status of the job (waiting, running) | ||
| + | * Complex possibilities for assigning priorities and deadlines to jobs, managing multiple queues etc. | ||
| + | * Ideally all computers in the cluster share the same environment and filesystem | ||
| + | * We have a simple training cluster for this exercise: | ||
| + | ** You submit jobs to queue on vyuka | ||
| + | ** They will run on computer cpu02 with 8 CPUs | ||
| + | ** This cluster is only temporarily available until next Wednesday | ||
| + | |||
| + | ===Submitting a job (qsub)=== | ||
| + | * <tt>qsub -b y -cwd 'command < input > output 2> error'</tt> | ||
| + | ** quoting around command allows us to include special characters, such as <, > etc. and not to apply it to qsub command itself | ||
| + | ** -b y treats command as binary, usually preferable for both binary programs and scripts | ||
| + | ** -cwd executes command in the current directory | ||
| + | ** -N name allows to set name of the job | ||
| + | ** -l resource=value requests some non-default resources | ||
| + | ** for example, we can use <tt>-l threads=2</tt> to request 2 threads for parallel programs | ||
| + | ** Grid engine will not check if you do not use more CPUs or memory than requested, be considerate (and perhaps occassionally watch your jobs by running top at the computer where they execute) | ||
| + | * qsub will create files for stdout and stderr, e.g. s2.o27 and s2.e27 for the job with name s2 and jobid 27 | ||
| + | |||
| + | ===Monitoring and deleting jobs (qstat, qdel)=== | ||
| + | * <tt>qstat</tt> displays jobs of the current user | ||
| + | <pre> | ||
| + | job-ID prior name user state submit/start at queue slots ja-task-ID | ||
| + | ----------------------------------------------------------------------------------------------------------------- | ||
| + | 28 0.50000 s3 bbrejova r 03/15/2016 22:12:18 main.q@cpu02.compbio.fmph.unib 1 | ||
| + | 29 0.00000 s3 bbrejova qw 03/15/2016 22:14:08 1 | ||
| + | </pre> | ||
| + | |||
| + | * <tt>qstat -u '*'</tt> displays jobs of all users | ||
| + | ** finished jobs disappear from the list | ||
| + | * <tt>qstat -F threads</tt> shows how many threads available | ||
| + | <pre> | ||
| + | queuename qtype resv/used/tot. load_avg arch states | ||
| + | --------------------------------------------------------------------------------- | ||
| + | main.q@cpu02.compbio.fmph.unib BIP 0/2/8 0.03 lx26-amd64 | ||
| + | hc:threads=0 | ||
| + | 28 0.75000 s3 bbrejova r 03/15/2016 22:12:18 1 | ||
| + | 29 0.25000 s3 bbrejova r 03/15/2016 22:14:18 1 | ||
| + | </pre> | ||
| + | |||
| + | * Command qdel allows you to delete a job (waiting or running) | ||
| + | |||
| + | ===Interactive work on the cluster (qrsh), screen=== | ||
| + | * qrsh creates a job which is a normal interactive shell running on the cluster | ||
| + | * in this shell you can then manually run commands | ||
| + | * however, when you close the shell, the job finishes | ||
| + | * therefore it is a good idea to run qrsh within screen | ||
| + | ** run screen command, this creates a new shell | ||
| + | ** within this shell, run qrsh, then whatever commands | ||
| + | ** by pressing Ctrl-a d you "detach" the screen, so that both shells (local and qrsh) continue running but you can close your local window | ||
| + | ** later by running screen -r you get back to your shells | ||
| + | |||
| + | ===Running many small jobs=== | ||
| + | For example tens of thousands of genes, runs some computation for each gene | ||
| + | * Have a script which iterates through all and runs them sequentially (as in HW02). | ||
| + | ** Problems: Does not use parallelism, needs more programming to restart after some interruption | ||
| + | * Submit processing of each gene as a separate job to cluster (submitting done by a script/one-liner). | ||
| + | ** Jobs can run in parallel on many different computers | ||
| + | ** Problem: Queue gets very long, hard to monitor progress, hard to resubmit only unfinished jobs after some failure. | ||
| + | * Array jobs in qsub (option -t): runs jobs numbered 1,2,3...; number of the job in an environment variable, used by the script to decide which gene to process | ||
| + | ** Queue contains only running sub-jobs plus one line for the remaining part of the array job. | ||
| + | ** After failure, you can resubmit only unfinished portion of the interval (e.g. start from job 173). | ||
| + | * Next: using make in which you specify how to process each gene and submit a single make command to the queue | ||
| + | ** Make can execute multiple tasks in parallel using several threads on the same computer (qsub array jobs can run tasks on multiple computers) | ||
| + | ** It will automatically skip tasks which are already finished | ||
| + | |||
| + | ==Make== | ||
| + | * [https://en.wikipedia.org/wiki/Make_(software) Make] is a system for automatically building programs (running compiler, linker etc) | ||
| + | ** In particular, we will use [https://www.gnu.org/software/make/manual/ GNU make] | ||
| + | * Rules for compilation are written in a Makefile | ||
| + | * Rather complex syntax with many features, we will only cover basics | ||
| + | |||
| + | ===Rules=== | ||
| + | * The main part of a Makefile are rules specifying how to generate target files from some source files (prerequisites). | ||
| + | * For example the following rule generates target.txt by concatenating source1.txt a source2.txt: | ||
| + | <pre> | ||
| + | target.txt : source1.txt source2.txt | ||
| + | cat source1.txt source2.txt > target.txt | ||
| + | </pre> | ||
| + | * The first line describes target and prerequisites, starts in the first column | ||
| + | * The following lines list commands to execute to create the target | ||
| + | * Each line with a command starts with a '''tab''' character | ||
| + | |||
| + | * If we now have a directory with this rule in Makefile and files source1.txt and source2.txt, running <tt>make target.txt</tt> will run the cat command | ||
| + | * However, if <tt>target.txt</tt> already exists, the command will be run only if one of the prerequisites has more recent modification time than the target | ||
| + | * This allows to restart interrupted computations or rerun necessary parts after modification of some input files | ||
| + | * Makefile automatically chains the rules as necessary: | ||
| + | ** if we run <tt>make target.txt</tt> and some prerequisite does not exist, Makefile checks if it can be created by some other rule and runs that rule first | ||
| + | ** In general it first finds all necessary steps and runs them in topological order so that each rules has its prerequisites ready | ||
| + | ** Option <tt>make -n target</tt> will show what commands would be executed to build target (dry run) - good idea before running something potentially dangerous | ||
| + | |||
| + | ===Pattern rules=== | ||
| + | |||
| + | * We can specify a general rule for files with a systematic naming scheme. For example, to create a .pdf file from a .tex file, we use pdflatex command: | ||
| + | <pre> | ||
| + | %.pdf : %.tex | ||
| + | pdflatex $^ | ||
| + | </pre> | ||
| + | * In the first line, % denotes some variable part of the filename, which has to agree in the target and all prerequisites | ||
| + | * In commands, we can use several variables: | ||
| + | ** $^ contains name for the prerequisite (source) | ||
| + | ** $@ contains the name of the target | ||
| + | ** $* contains the string matched by % | ||
| + | |||
| + | ===Other useful tricks in Makefiles=== | ||
| + | |||
| + | ====Variables==== | ||
| + | * Store some reusable values in variables, then use them several times in the Makefile: | ||
| + | <pre> | ||
| + | PATH := /projects/trees/bin | ||
| + | |||
| + | target : source | ||
| + | $(PATH)/script < $^ > $@ | ||
| + | </pre> | ||
| + | |||
| + | ====Wildcards, creating a list of targets from files in the directory==== | ||
| + | |||
| + | The following Makefile automatically creates .png version of each .eps file simply by running make: | ||
| + | <pre> | ||
| + | EPS := $(wildcard *.eps) | ||
| + | EPSPNG := $(patsubst %.eps,%.png,$(EPS)) | ||
| + | |||
| + | all: $(EPSPNG) | ||
| + | |||
| + | clean: | ||
| + | rm $(EPSPNG) | ||
| + | |||
| + | %.png : %.eps | ||
| + | convert -density 250 $^ $@ | ||
| + | </pre> | ||
| + | * variable EPS contains names of all files matching *.eps | ||
| + | * variable EPSPNG contains desirable names of png files | ||
| + | ** it is created by taking filenams in EPS and changing.eps to .png | ||
| + | * <tt>all</tt> if a "phony target" which is not really created | ||
| + | ** its rule has no commands but all png files are prerequisites, so are done first | ||
| + | ** the first target in Makefile (in this case <tt>all</tt>) is default when no other target specified on command-line | ||
| + | * <tt>clean</tt> is also phony target for deleting generated png files | ||
| + | |||
| + | ====Useful special built-in target names==== | ||
| + | Include these lines in your Makefile if desired | ||
| + | <pre> | ||
| + | .SECONDARY: | ||
| + | # prevents deletion of intermediate targets in chained rules | ||
| + | |||
| + | .DELETE_ON_ERROR: | ||
| + | # delete targets if a rule fails | ||
| + | </pre> | ||
| + | |||
| + | ===Parallel make=== | ||
| + | * running make with option <tt>-j 4</tt> will run up to 4 commands in parallel if their dependencies are already finished | ||
| + | * easy parallelization on a single computer | ||
| + | |||
| + | ==Alternatives to Makefiles== | ||
| + | * Bioinformatics often uses "pipelines" - sequences of commands run one after another, e.g. by a script of Makefile | ||
| + | * There are many tools developed for automating computational pipelines, see e.g. this review: [https://academic.oup.com/bib/article/doi/10.1093/bib/bbw020/2562749/A-review-of-bioinformatic-pipeline-frameworks Jeremy Leipzig; A review of bioinformatic pipeline frameworks. Brief Bioinform 2016 bbw020.] | ||
| + | * For example [https://bitbucket.org/snakemake/snakemake/wiki/Home Snakemake] | ||
| + | ** Workflows can contain shell commands or Python code | ||
| + | ** Big advantage compared to Make: pattern rules my contain multiple variable portions (in make only one % per filename) | ||
| + | ** For example, you have several fasta files and several HMMs representing protein families and you wans to run each HMM on each fasta file: | ||
| + | <pre> | ||
| + | rule HMMER: | ||
| + | input: "{filename}.fasta", "{hmm}.hmm" | ||
| + | output: "{filename}_{hmm}.hmmer" | ||
| + | shell: "hmmsearch --domE 1e-5 --noali --domtblout {output} {input[1]} {input[0]}" | ||
| + | </pre> | ||
| + | =HW04= | ||
| + | See also [[#L02|Lecture 2]], [[#HW02]] | ||
| + | |||
| + | In this homework, we will return to the example in [[#HW02|homework 2]], where we took genes from several organisms, found orthogroups of corresponding genes and built a phylogenetic tree for each orthogroup. This was all done in a single big Perl script. In this homework, we will write a similar pipeline using make and execute it remotely using qsub. We will use proteins instead of DNA and we will use a different set of species. Most of the work is already done, only small modifications are necessary. | ||
| + | |||
| + | * Submit by copying requested files to /submit/hw04/username/ | ||
| + | * Do not forget to submit protocol, outline of the protocol is in /tasks/hw04/protocol.txt | ||
| + | |||
| + | ==Task A== | ||
| + | |||
| + | * In this task, you will run a long alignment job (>1 hour) | ||
| + | * Copy directory /tasks/hw04/large to your home directory | ||
| + | ** ref.fa: all proteins from yeast ''Yarrowia lipolytica'' | ||
| + | ** other.fa: all proteins from 8 other yeast species | ||
| + | ** Makefile: run blast on ref.fa vs other.fa (also formats database other.fa before that) | ||
| + | * run make -n to see what commands will be done (you should see formatdb and blastall + echo for timing), copy the output to the '''protocol''' | ||
| + | * run qsub with appropriate options to run make (at least -cwd and -b y) | ||
| + | * then run qstat > queue.txt | ||
| + | ** '''Submit''' file queue.txt showing your job waiting or running | ||
| + | * When your job finishes, '''submit''' also the following two files: | ||
| + | ** the last 100 lines from the output file ref.blast under the name ref-end.blast (use tool tail -n 100) | ||
| + | ** standard output from the qsub job, which is stored in a file named e.g. make.oX where X is the number of your job. The output shows the time when your job started and finished (this information was written by commands echo in the Makefile) | ||
| + | |||
| + | ==Task B== | ||
| + | |||
| + | * In this task, you will finish a Makefile for splitting blast results into orthogroups and building phylogenetic trees for each group | ||
| + | ** This Makefile works with much smaller files and so you can run it many times on vyuka, without qsub | ||
| + | ** If it runs too slowly, you can temporarily modify ref.fa to contain only the first 3 sequences, debug your makefile and then again copy the original ref.fa from /tasks/hw04/small to run the final analysis | ||
| + | * Copy directory /tasks/hw04/small to your home directory | ||
| + | ** ref.fa: 10 proteins from yeast ''Yarrowia lipolytica'' | ||
| + | ** other.fa: a selected subset of proteins from 8 other yeast species | ||
| + | ** Makefile: longer makefile | ||
| + | |||
| + | The Makefile runs the analysis in four stages. Stages 1,2 and 4 are done, you have to finish stage 3 | ||
| + | * If you run make without argument, it will attempt to run all 4 stages, but stage 3 will not run, because it is missing | ||
| + | * Stage 1: run as <tt>make ref.brm</tt> | ||
| + | ** It runs blast as in task A, then splits proteins into orthogroups and creates one directory for each group with file prot.fa containing protein sequences | ||
| + | * Stage 2: run as <tt>make alignments</tt> | ||
| + | ** In each directory with a single gene, it will create an alignment prot.phy and link it under names lg.phy and wag.phy | ||
| + | * Stage 3: run as <tt>make trees</tt> (needs to be written by you) | ||
| + | ** In each directory with a single gene, it should create lg.phy_phyml_tree and wag.phy_phyml_tree | ||
| + | ** These corresponds to results of phyml commands run with two different evolutionary models WAG and LG, where LG is the default | ||
| + | ** Run phyml by commands of the forms: | ||
| + | *** <tt>phyml -i INPUT --datatype aa --bootstrap 0 --no_memory_check >LOG</tt> | ||
| + | *** <tt>phyml -i INPUT --model WAG --datatype aa --bootstrap 0 --no_memory_check >LOG</tt> | ||
| + | ** Change INPUT and LOG in the commands to appropriate filenames using make variables $@, $^, $* etc. Input should come from lg.phy or wag.phy in the directory of a gene and log should be the same as tree name with extension .log added (e.g. lg.phy_phyml_tree.log) | ||
| + | ** Also add variables LG_TREES and WAG_TREES listing filenames of all desirable trees and uncomment phony target trees wich uses these variables | ||
| + | * Stage 4: run as <tt>make consensus</tt> | ||
| + | ** Output trees from stage 3 are concatenated for each model separately to files lg/intree wag/intree and then phylip is run to produce consensus trees lg.tree and wag.tree | ||
| + | ** This stage also needs variables LG_TREES and WAG_TREES to be defined by you. | ||
| + | |||
| + | * Run your Makefile | ||
| + | * '''Submit''' the whole directory small, including Makefile and all gene directories with tree files. | ||
| + | * Look at the two resulting trees using the figtree program, switch on displaying branch labels in the left panel with options. These labels show for each branch of the tree, how many of the input trees support this branch. '''Write to your protocol''', what do you observe: Do the two trees differ? If yes, do they differ in branches supported by many different genes trees, or few? What is the highest and lowest support for a branch in each tree? | ||
| + | |||
| + | ==Further possibilities== | ||
| + | |||
| + | Here are some possibilities for further experiments, in case you are interested (do not submit these): | ||
| + | * You could copy your extended Makefile to directory large and create trees for all orthogroups in the big set | ||
| + | ** This would take a long time, so submit it through qsub and only some time after the lecture is over to allow classmates to work on task A | ||
| + | ** After ref.brm si done, programs for individual genes can be run in parallel, so you can try running make -j 2 and request 2 threads from qsub | ||
| + | * Phyml also supports other models, for example JTT (see [http://www.atgc-montpellier.fr/download/papers/phyml_manual_2012.pdf manual]), you could try to play with those. | ||
| + | * Command touch FILENAME will change modification time of the given file to current file | ||
| + | ** What happens when you run touch on some of the intermediate files in the analysis in task B? Does Makefile always run properly? | ||
| + | =L05= | ||
| + | [[#HW05]] | ||
| + | |||
| + | Program for today: basics of R (applied to biology examples) | ||
| + | * very short intro as a lecture | ||
| + | * tutorial as HW: read a bit of text, try some commands, extend/modify them as requested | ||
| + | |||
| + | In this course we cover several languages popular for scripting in bioinformatics: Perl, Python, R | ||
| + | * their capabilities overlap, many extensions emulate strengths of one in another | ||
| + | * choose a language based on your preference, level of knowledge, existing code for the task, rest of the team | ||
| + | * quickly learn a new language if needed | ||
| + | * also possibly combine, e.g. preprocess data in Perl or Python, then run statistical analyses in R, automate entire pipeline with bash or make | ||
| + | |||
| + | ==Introduction== | ||
| + | * [http://www.r-project.org/ R] is an open-source system for statistical computing and data visualization | ||
| + | * Programming language, command-line interface | ||
| + | * Many built-in functions, additional libraries | ||
| + | ** For example http://bioconductor.org/ for bioinformatics | ||
| + | * We will concentrate on useful commands rather than language features | ||
| + | |||
| + | ==Working in R== | ||
| + | * Run command R, type commands in command-line interface | ||
| + | ** supports history of commands (arrows, up and down, Ctrl-R) and completing command names with tab key | ||
| + | <pre> | ||
| + | > 1+2 | ||
| + | [1] 3 | ||
| + | </pre> | ||
| + | * Write a script to file, run it from command-line: <tt>R --vanilla --slave < file.R</tt> | ||
| + | * Use <tt>rstudio</tt> to open a graphics IDE [https://www.rstudio.com/products/RStudio/] | ||
| + | ** Windows with editor of R scripts, console, variables, plots | ||
| + | ** Ctrl-Enter in editor executes current command in console | ||
| + | <pre> | ||
| + | x=c(1:10) | ||
| + | plot(x,x*x) | ||
| + | </pre> | ||
| + | * <tt>? plot</tt> displays help for plot command | ||
| + | |||
| + | Suggested workflow | ||
| + | * work interactively in Rstudio or on command line, try various options | ||
| + | * select useful commands, store in a script | ||
| + | * run script automatically on new data/new versions, potentially as a part of a bigger pipeline | ||
| + | |||
| + | ==Additional information== | ||
| + | * [http://cran.r-project.org/doc/manuals/R-intro.html Official tutorial] | ||
| + | * [http://cran.r-project.org/doc/contrib/Seefeld_StatsRBio.pdf Seefeld, Linder: Statistics Using R with Biological Examples (pdf book)] | ||
| + | * [http://www.burns-stat.com/pages/Tutor/R_inferno.pdf Patrick Burns: The R Inferno] (intricacies of the language) | ||
| + | * [https://www.r-project.org/doc/bib/R-books.html Other books] | ||
| + | |||
| + | ==Gene expression data== | ||
| + | * Gene expression: DNA->mRNA->protein | ||
| + | * Level of gene expression: Extract mRNA from a cell, measure amounts of mRNA | ||
| + | * Technologies: microarray, RNA-seq | ||
| + | Gene expression data | ||
| + | * Rows: genes | ||
| + | * Columns: experiments (e.g. different conditions or different individuals) | ||
| + | * Each value is expression of a gene, i.e. relative amount of mRNA for this gene in the sample | ||
| + | |||
| + | We will use microarray data for yeast: | ||
| + | * Strassburg, Katrin, et al. "Dynamic transcriptional and metabolic responses in yeast adapting to temperature stress." Omics: a journal of integrative biology 14.3 (2010): 249-259. [http://online.liebertpub.com/doi/full/10.1089/omi.2009.0107] | ||
| + | * Downloaded from GEO database [http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE15352] | ||
| + | * Data already preprocessed: normalization, log2, etc | ||
| + | * We have selected only cold conditions, genes with absolute change at least 1 | ||
| + | * Data: 2738 genes, 8 experiments in a time series, yeast moved from normal temperature 28 degrees C to cold conditions 10 degrees C, samples taken after 0min, 15min, 30min, 1h, 2h, 4h, 8h, 24h in cold | ||
| + | =HW05= | ||
| + | [[#L05]] | ||
| + | |||
| + | In this homework, try to read text, execute given commands, potentially trying some small modifications. | ||
| + | * Then do tasks A-D, submit required files (3x .png) | ||
| + | * In your protocol, enter commands used in tasks A-D, with explanatory comments in more complicated situations | ||
| + | * In task B also enter required output to protocol | ||
| + | |||
| + | ==First steps== | ||
| + | * Type a command, R writes the answer, e.g.: | ||
| + | <pre> | ||
| + | > 1+2 | ||
| + | [1] 3 | ||
| + | </pre> | ||
| + | * We can store values in variables and use them later on | ||
| + | <pre> | ||
| + | > # The size of the sequenced portion of cow's genome, in millions of base pairs | ||
| + | > Cow_genome_size <- 2290 | ||
| + | > Cow_genome_size | ||
| + | [1] 2290 | ||
| + | > Cow_chromosome_pairs <- 30 | ||
| + | > Cow_avg_chrom <- Cow_genome_size / Cow_chromosome_pairs | ||
| + | > Cow_avg_chrom | ||
| + | [1] 76.33333 | ||
| + | </pre> | ||
| + | Surprises: | ||
| + | * dots are used as parts of id's, e.g. read.table is name of a single function (not method for object read) | ||
| + | * assignment via <- or = | ||
| + | ** careful: a<-3 is assignment, a < -3 is comparison | ||
| + | * vectors etc are indexed from 1, not from 0 | ||
| + | |||
| + | ==Vectors, basic plots== | ||
| + | * Vector is a sequence of values of the same type (all are numbers or all are strings or all are booleans) | ||
| + | <pre> | ||
| + | # Vector can be created from a list of numbers by function c | ||
| + | a<-c(1,2,4) | ||
| + | a | ||
| + | # prints [1] 1 2 4 | ||
| + | |||
| + | # function c also concatenates vectors | ||
| + | c(a,a) | ||
| + | # prints [1] 1 2 4 1 2 4 | ||
| + | |||
| + | # Vector of two strings | ||
| + | b<-c("hello", "world") | ||
| + | |||
| + | # Create a vector of numbers 1..10 | ||
| + | x<-1:10 | ||
| + | x | ||
| + | # prints [1] 1 2 3 4 5 6 7 8 9 10 | ||
| + | </pre> | ||
| + | |||
| + | ===Vector arithmetics=== | ||
| + | * Operations applied to each member of the vector | ||
| + | <pre> | ||
| + | x<-1:10 | ||
| + | # Square each number in vector x | ||
| + | x*x | ||
| + | # prints [1] 1 4 9 16 25 36 49 64 81 100 | ||
| + | |||
| + | # New vector y: logarithm from a number in x squared | ||
| + | y<-log(x*x) | ||
| + | y | ||
| + | # prints [1] 0.000000 1.386294 2.197225 2.772589 3.218876 3.583519 3.891820 4.158883 | ||
| + | # [9] 4.394449 4.605170 | ||
| + | |||
| + | # Draw graph of function log(x*x) for x=1..10 | ||
| + | plot(x,y) | ||
| + | # The same graph but use lines instead of dots | ||
| + | plot(x,y,type="l") | ||
| + | |||
| + | # Addressing elements of a vector: positions start at 1 | ||
| + | # Second element of the vector | ||
| + | y[2] | ||
| + | # prints [1] 1.386294 | ||
| + | |||
| + | # Which elements of the vector satisfy certain condition? (vector of logical values) | ||
| + | y>3 | ||
| + | # prints [1] FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE | ||
| + | |||
| + | # write only those elements from y that satisfy the condition | ||
| + | y[y>3] | ||
| + | # prints [1] 3.218876 3.583519 3.891820 4.158883 4.394449 4.605170 | ||
| + | |||
| + | # we can also write values of x such that values of y satisfy the condition... | ||
| + | x[y>3] | ||
| + | # prints [1] 5 6 7 8 9 10 | ||
| + | </pre> | ||
| + | |||
| + | * Alternative plotting facilities: [http://ggplot2.org/ ggplot2 library], [https://cran.r-project.org/web/packages/lattice/index.html lattice library] | ||
| + | |||
| + | ===Task A=== | ||
| + | * Create a plot of the binary logarithm with dots in the graph more densely spaced (from 0.1 to 10 with step 0.1) | ||
| + | * Store it in file <tt>log.png</tt> and '''submit''' this file | ||
| + | * Hints: | ||
| + | ** Create x and y by vector arithmetics | ||
| + | ** To compute binary logarithm check help <tt>? log</tt> | ||
| + | ** Before running plot, use command <tt>png("log.png")</tt> to store the result, afterwards call dev.off() to close the file (in rstudio you can also export plots manually) | ||
| + | |||
| + | ==Data frames and simple statistics== | ||
| + | * Data frame: table similar to spreadsheet, each column is a vector, all are of the same length | ||
| + | * We will use a table with the following columns: | ||
| + | ** The size of the sequenced portion of a genome, in millions of base pairs | ||
| + | ** Number of chromosome pairs | ||
| + | ** GC content | ||
| + | ** taxonomic group mammal or fish | ||
| + | * Stored in CSV format, columns separated by tabs. | ||
| + | * Data: Han et al Genome Biology 2008 [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2441465/] | ||
| + | <pre> | ||
| + | Species Size Chrom GC Group | ||
| + | Human 2850 23 40.9 mammal | ||
| + | Chimpanzee 2750 24 40.7 mammal | ||
| + | Macaque 2650 21 40.7 mammal | ||
| + | Mouse 2480 20 41.7 mammal | ||
| + | ... | ||
| + | Tetraodon 187 21 45.9 fish | ||
| + | ... | ||
| + | </pre> | ||
| + | |||
| + | <pre> | ||
| + | # reading a frame from file | ||
| + | a<-read.table("/tasks/hw05/genomes.csv", header = TRUE, sep = "\t"); | ||
| + | # column with name size | ||
| + | a$Size | ||
| + | |||
| + | # Average chromosome length: divide size by the number of chromosomes | ||
| + | a$Size/a$Chrom | ||
| + | |||
| + | # Add average chromosome length as a new column to frame a | ||
| + | a<-cbind(a,AvgChrom=a$Size/a$Chrom) | ||
| + | |||
| + | # scatter plot average chromosome length vs GC content | ||
| + | plot(a$AvgChrom, a$GC) | ||
| + | |||
| + | # compactly display structure of a | ||
| + | # (good for checking that import worked etc) | ||
| + | str(a) | ||
| + | |||
| + | # display mean, median, etc. of each column | ||
| + | summary(a); | ||
| + | |||
| + | # average genome size | ||
| + | mean(a$Size) | ||
| + | # average genome size for mammals | ||
| + | mean(a$Size[a$Group=="mammal"]) | ||
| + | # Standard deviation | ||
| + | sd(a$Size) | ||
| + | |||
| + | # Histogram of genome sizes | ||
| + | hist(a$Size) | ||
| + | </pre> | ||
| + | |||
| + | ===Task B=== | ||
| + | * Divide frame <tt>a</tt> to two frames, one for mammals, one for fish. Hint: | ||
| + | ** Try command <tt>a[c(1,2,3),]</tt>. What is it doing? | ||
| + | ** Try command <tt>a$Group=="mammal"</tt>. | ||
| + | ** Combine these two commands to get rows for all mammals and store the frame in a new variable, then repeat for fish | ||
| + | ** Use a general approach which does not depend on the exact number and ordering of rows in the table. | ||
| + | |||
| + | * Run the command <tt>summary</tt> separately for mammals and for fish. Which of their characteristics are different? | ||
| + | ** '''Write''' output and your conclusion to the protocol | ||
| + | |||
| + | ===Task C=== | ||
| + | * Draw a graph comparing genome size vs GC content; use different colors for points representing mammals and fish | ||
| + | ** '''Submit''' the plot in file <tt>genomes.png</tt> | ||
| + | ** To draw the graph, you can use one of the options below, or find yet another way | ||
| + | ** Option 1: first draw mammals with one color, then add fish in another color | ||
| + | *** Color of points can be changed by: <tt>plot(1:10,1:10, col="red")</tt> | ||
| + | *** After plot command you can add more points to the same graph by command <tt>points</tt>, which can be used similarly as <tt>plot</tt> | ||
| + | *** Warning: command <tt>points</tt> does not change the ranges of x and y axes. You have to set these manually so that points from both groups are visible. You can do this using options <tt>xlim</tt> and <tt>ylim</tt>, e.g. <tt>plot(x,y, col="red", xlim=c(1,100), ylim=c(1,100))</tt> | ||
| + | ** Option 2: plot both mammals and fish in one plot command, and give it a vector of colors, one for each point | ||
| + | *** <tt>plot(1:10,1:10,col=c(rep("red",5),rep("blue",5)))</tt> will plot the first 5 points red and the last 5 points blue | ||
| + | |||
| + | * Bonus task: add a legend to the plot, showing which color is mammal and which is fish | ||
| + | |||
| + | ==Expression data and clustering== | ||
| + | |||
| + | Data here is bigger, better to use plain R rather than rstudio (limited server CPU/memory) | ||
| + | |||
| + | <pre> | ||
| + | # Read gene expression data table | ||
| + | a<-read.table("/tasks/hw05/microarray.csv", header = TRUE, sep = "\t", row.names=1) | ||
| + | # Visual check of the first row | ||
| + | a[1,] | ||
| + | # plot starting point vs. situation after 1 hour | ||
| + | plot(a$cold_0min,a$cold_1h) | ||
| + | # to better see density in dense clouds of points, use this plot | ||
| + | smoothScatter(a$cold_15min,a$cold_1h) | ||
| + | # outliers away from diagonal in the plot above are most strongly differentially expressed genes | ||
| + | # these are more easy to see in MA plot: | ||
| + | # x-axis: average expression in the two conditions | ||
| + | # y-axis: difference between values (they are log-scale, so difference 1 means 2-fold) | ||
| + | smoothScatter((a$cold_15min+a$cold_1h)/2,a$cold_15min-a$cold_1h) | ||
| + | </pre> | ||
| + | |||
| + | Clustering is a wide group of methods that split data points into groups with similar properties | ||
| + | * We will group together genes that have a similar reaction to cold, i.e. their rows in gene expression data matrix have similar values | ||
| + | We will consider two simple clustering methods | ||
| + | * K means clustering splits points (genes) into k clusters, where k is a parameter given by the user. It finds a center of each cluster and tries to minimize the sum of distances from individual points to the center of their cluster. Note that this algorithm is randomized so you will get different clusters each time. | ||
| + | * Hierarchical clustering puts all data points (genes) to a hierarchy so that smallest subtrees of the hierarchy are the most closely related groups of points and these are connected to bigger and more loosely related groups. | ||
| + | |||
| + | [[Image:HW08-heatmap.png|thumb|200px|right|Example of a heatmap]] | ||
| + | <pre> | ||
| + | # Heatmap: creates hierarchical clustering of rows | ||
| + | # then shows every value in the table using color ranging from red (lowest) to white (highest) | ||
| + | # Computation may take some time | ||
| + | heatmap(as.matrix(a),Colv=NA) | ||
| + | # Previous heatmap normalized each row, the next one uses data as they are: | ||
| + | heatmap(as.matrix(a),Colv=NA,scale="none") | ||
| + | </pre> | ||
| + | |||
| + | <pre> | ||
| + | # k means clustering to 7 clusters | ||
| + | k=7 | ||
| + | cl <- kmeans(a,k) | ||
| + | # each gene has assigned a cluster (number between 1 and k) | ||
| + | cl$cluster | ||
| + | # draw only cluster number 3 out of k | ||
| + | heatmap(as.matrix(a[cl$cluster==3,]),Rowv=NA, Colv=NA) | ||
| + | |||
| + | # reorder genes in the table according to cluster | ||
| + | heatmap(as.matrix(a[order(cl$cluster),]),Rowv=NA, Colv=NA) | ||
| + | |||
| + | # compare overall column means with column means in cluster 3 | ||
| + | # function apply uses mean on every column (or row if 2 changed to 1) | ||
| + | apply(a,2,mean) | ||
| + | # now means within cluster | ||
| + | apply(a[cl$cluster==3,],2,mean) | ||
| + | |||
| + | # clusters have centers which are also computed as means | ||
| + | # so this is the same as previous command | ||
| + | cl$centers[3,] | ||
| + | </pre> | ||
| + | |||
| + | ===Task D=== | ||
| + | [[Image:HW08-clusters.png|thumb|200px|right|Example of a required plot]] | ||
| + | * Draw a plot in which x-axis is time and y-axis is the expression level and the center of each cluster is shown as a line | ||
| + | ** use command <tt>matplot(x,y,type="l")</tt> which gets two matrices x and y and plots columns of one vs the other | ||
| + | ** <tt>matplot(,y,type="l")</tt> will use numbers 1,2,3... as columns of the missing matrix x | ||
| + | ** create y from <tt>cl$centers</tt> by applying function <tt>t</tt> (transpose) | ||
| + | ** to create an appropriate matrix x, create a vector of times for individual experiments in minutes or hours (do it manually, no need to parse column names automatically) | ||
| + | ** using functions <tt>rep</tt> and <tt>matrix</tt> you can create a matrix x in which this vector is used as every column | ||
| + | ** then run <tt>matplot(x,y,type="l")</tt> | ||
| + | ** since time points are not evenly spaced, it would be better to use logscale: <tt>matplot(x,y,type="l",log="x")</tt> | ||
| + | ** to avoid log(0), change the first timepoint from 0min to 1min | ||
| + | * Submit file '''clusters.png''' with your final plot | ||
| + | =L06= | ||
| + | [[#HW06]] | ||
| + | |||
| + | Topic of this lecture are statistical tests in R. | ||
| + | * Beginners in statistics: listen to lecture, then do tasks A, B, C | ||
| + | * If you know basics of statistical tests, do tasks B, C, D | ||
| + | * More information on this topic in [https://sluzby.fmph.uniba.sk/infolist/sk/1-EFM-340_13.html 1-EFM-340 Počítačová štatistika] | ||
| + | |||
| + | ==Introduction to statistical tests: sign test== | ||
| + | * [https://en.wikipedia.org/wiki/Sign_test] | ||
| + | * Two friends ''A'' and ''B'' have played their favourite game ''n''=10 times, ''A'' has won 6 times and ''B'' has won 4 times. | ||
| + | * ''A'' claims that he is a better player, ''B'' claims that such a result could easily happen by chance if they were equally good players. | ||
| + | * Hypothesis of player ''B'' is called ''null hypothesis'' that the pattern we see (''A'' won more often than ''B'') is simply a result of chance | ||
| + | * Null hypothesis reformulated: we toss coin ''n'' times and compute value ''X'': the number of times we see head. The tosses are independent and each toss has equal probability of being 0 or 1 | ||
| + | * Similar situation: comparing programs A and B on several inputs, counting how many times is program A better than B. | ||
| + | <pre> | ||
| + | # simulation in R: generate 10 psedorandom bits | ||
| + | # (1=player A won) | ||
| + | sample(c(0,1), 10, replace = TRUE) | ||
| + | # result e.g. 0 0 0 0 1 0 1 1 0 0 | ||
| + | |||
| + | # directly compute random variable X, i.e. sum of bits | ||
| + | sum(sample(c(0,1), 10, replace = TRUE)) | ||
| + | # result e.g. 5 | ||
| + | |||
| + | # we define a function which will m times repeat | ||
| + | # the coin tossing experiment with n tosses | ||
| + | # and returns a vector with m values of random variable X | ||
| + | experiment <- function(m, n) { | ||
| + | x = rep(0, m) # create vector with m zeroes | ||
| + | for(i in 1:m) { # for loop through m experiments | ||
| + | x[i] = sum(sample(c(0,1), n, replace = TRUE)) | ||
| + | } | ||
| + | return(x) # return array of values | ||
| + | } | ||
| + | # call the function for m=20 experiments, each with n tosses | ||
| + | experiment(20,10) | ||
| + | # result e.g. 4 5 3 6 2 3 5 5 3 4 5 5 6 6 6 5 6 6 6 4 | ||
| + | # draw histograms for 20 experiments and 1000 experiments | ||
| + | png("hist10.png") # open png file | ||
| + | par(mfrow=c(2,1)) # matrix of plots with 2 rows and 1 column | ||
| + | hist(experiment(20,10)) | ||
| + | hist(experiment(1000,10)) | ||
| + | dev.off() # finish writing to file | ||
| + | </pre> | ||
| + | * It is easy to realize that we get [https://en.wikipedia.org/wiki/Binomial_distribution binomial distribution] (binomické rozdelenie) | ||
| + | * <math>Pr(X=k) = {n \choose k} 2^{-n}</math> | ||
| + | * ''P-value'' of the test is the probability that simply by chance we would get ''k'' the same or more extreme than in our data. | ||
| + | * In other words, what is the probability that in 10 tosses we see head 6 times or more (one sided test) | ||
| + | * <math>\sum_{j=k}^n {n \choose k} 2^{-n}</math> | ||
| + | * If the p-value is very small, say smaller than 0.01, we reject the null hypothesis and assume that player ''A'' is in fact better than ''B'' | ||
| + | |||
| + | <pre> | ||
| + | # computing the probability that we get exactly 6 heads in 10 tosses | ||
| + | dbinom(6, 10, 0.5) # result 0.2050781 | ||
| + | # we get the same as our formula above: | ||
| + | 7*8*9*10/(2*3*4*(2^10)) # result 0.2050781 | ||
| + | |||
| + | # entire probability distribution: probabilities 0..10 heads in 10 tosses | ||
| + | dbinom(0:10, 10, 0.5) | ||
| + | # [1] 0.0009765625 0.0097656250 0.0439453125 0.1171875000 0.2050781250 | ||
| + | # [6] 0.2460937500 0.2050781250 0.1171875000 0.0439453125 0.0097656250 | ||
| + | # [11] 0.0009765625 | ||
| + | |||
| + | #we can also plot the distribution | ||
| + | plot(0:10, dbinom(0:10, 10, 0.5)) | ||
| + | barplot(dbinom(0:10,10,0.5)) | ||
| + | |||
| + | #our p-value is sum for 7,8,9,10 | ||
| + | sum(dbinom(6:10,10,0.5)) | ||
| + | # result: 0.3769531 | ||
| + | # so results this "extreme" are not rare by chance, | ||
| + | # they happen in about 38% of cases | ||
| + | |||
| + | # R can compute the sum for us using pbinom | ||
| + | # this considers all values greater than 5 | ||
| + | pbinom(5, 10, 0.5, lower.tail=FALSE) | ||
| + | # result again 0.3769531 | ||
| + | |||
| + | # if probability is too small, use log of it | ||
| + | pbinom(9999, 10000, 0.5, lower.tail=FALSE, log.p = TRUE) | ||
| + | # [1] -6931.472 | ||
| + | # the probability of getting 10000x head is exp(-6931.472) = 2^{-100000} | ||
| + | |||
| + | # generating numbers from binomial distribution | ||
| + | # - similarly to our function experiment | ||
| + | rbinom(20, 10, 0.5) | ||
| + | # [1] 4 4 8 2 6 6 3 5 5 5 5 6 6 2 7 6 4 6 6 5 | ||
| + | |||
| + | # running the test | ||
| + | binom.test(6, 10, p = 0.5, alternative="greater") | ||
| + | # | ||
| + | # Exact binomial test | ||
| + | # | ||
| + | # data: 6 and 10 | ||
| + | # number of successes = 6, number of trials = 10, p-value = 0.377 | ||
| + | # alternative hypothesis: true probability of success is greater than 0.5 | ||
| + | # 95 percent confidence interval: | ||
| + | # 0.3035372 1.0000000 | ||
| + | # sample estimates: | ||
| + | # probability of success | ||
| + | # 0.6 | ||
| + | |||
| + | # to only get p-value run | ||
| + | binom.test(6, 10, p = 0.5, alternative="greater")$p.value | ||
| + | # result 0.3769531 | ||
| + | </pre> | ||
| + | |||
| + | ==Comparing two sets of values: Welch's t-test== | ||
| + | * Let us now consider two sets of values drawn from two [https://en.wikipedia.org/wiki/Normal_distribution normal distributions] with unknown means and variances | ||
| + | * The null hypothesis of the [https://en.wikipedia.org/wiki/Welch%27s_t-test Welch's t-test] is that the two distributions have equal means | ||
| + | * The test computes test statistics (in R for vectors x1, x2): | ||
| + | ** <tt>(mean(x1)-mean(x2))/sqrt(var(x1)/length(x1)+var(x2)/length(x2))</tt> | ||
| + | * This test statistics is approximately distributed according to [https://en.wikipedia.org/wiki/Student%27s_t-distribution Student's t-distribution] with the degree of freedom obtained by | ||
| + | <pre> | ||
| + | n1=length(x1) | ||
| + | n2=length(x2) | ||
| + | (var(x1)/n1+var(x2)/n2)**2/(var(x1)**2/((n1-1)*n1*n1)+var(x2)**2/((n2-1)*n2*n2)) | ||
| + | </pre> | ||
| + | * Luckily R will compute the test for us simply by calling t.test | ||
| + | <pre> | ||
| + | x1 = rnorm(6, 2, 1) | ||
| + | # 2.70110750 3.45304366 -0.02696629 2.86020145 2.37496993 2.27073550 | ||
| + | |||
| + | x2 = rnorm(4, 3, 0.5) | ||
| + | # 3.258643 3.731206 2.868478 2.239788 | ||
| + | > t.test(x1,x2) | ||
| + | # t = -1.2898, df = 7.774, p-value = 0.2341 | ||
| + | # alternative hypothesis: true difference in means is not equal to 0 | ||
| + | # means 2.272182 3.024529 | ||
| + | |||
| + | x2 = rnorm(4, 5, 0.5) | ||
| + | # 4.882395 4.423485 4.646700 4.515626 | ||
| + | t.test(x1,x2) | ||
| + | # t = -4.684, df = 5.405, p-value = 0.004435 | ||
| + | # means 2.272182 4.617051 | ||
| + | |||
| + | # to get only p-value, run | ||
| + | t.test(x1,x2)$p.value | ||
| + | </pre> | ||
| + | |||
| + | We will apply Welch's t-test to microarray data | ||
| + | * Data from GEO database [http://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS2925], publication [http://femsyr.oxfordjournals.org/content/7/6/819.abstract] | ||
| + | * Abbott et al 2007: Generic and specific transcriptional responses to different weak organic acids in anaerobic chemostat cultures of Saccharomyces cerevisiae | ||
| + | * gene expression measurements under 5 conditions: | ||
| + | ** reference: yeast grown in normal environment | ||
| + | ** 4 different acids added so that cells grow 50% slower (acetic, propionic, sorbic, benzoic) | ||
| + | * from each condition (reference and each acid) we have 3 replicates | ||
| + | * together our table has 15 columns (3 replicates from 5 conditions) | ||
| + | * 6398 rows (genes) | ||
| + | * We will test statistical difference between the reference condition and one of the acids (3 numbers vs other 2 numbers) | ||
| + | * See Task B in [[#HW06]] | ||
| + | |||
| + | ==Multiple testing correction== | ||
| + | |||
| + | * When we run t-tests on the reference vs. acetic acid on all 6398 genes, we get 118 genes with p-value<=0.01 | ||
| + | * Purely by chance this would happen in 1% of cases (from definition of p-value) | ||
| + | * So purely by chance we would expect to get about 64 genes with p-value<=0.01 | ||
| + | * So perhaps roughly half of our detected genes (maybe less, maybe more) are false positives | ||
| + | * Sometimes false positives may even overwhelm results | ||
| + | * Multiple testing correction tries to limit the number of false positives among results of multiple statistical tests | ||
| + | * Many different methods | ||
| + | * The simplest one is [https://en.wikipedia.org/wiki/Bonferroni_correction Bonferroni correction], where the threshold on p-value is divided by the number of tested genes, so instead of 0.01 we use 0.01/6398 = 1.56e-6 | ||
| + | * This way the expected overall number of false positives in the whole set is 0.01 and so the probability of getting even a single false positive is also at most 0.01 (by Markov inequality) | ||
| + | * We could instead multiply all p-values by the number of tests and apply the original threshold 0.01 - such artificially modified p-values are called corrected | ||
| + | * After Bonferroni correction we get only 1 significant gene | ||
| + | <pre> | ||
| + | # the results of p-tests are in vector pa of length 6398 | ||
| + | # manually multiply p-values by length(pa), count those that have value <=0.01 | ||
| + | sum(pa * length(pa) < 0.01) | ||
| + | # in R you can use p.adjust form multiple testing correction | ||
| + | pa.adjusted = p.adjust(pa, method ="bonferroni") | ||
| + | # this is equivalent to multiplying by the length and using 1 if the result > 1 | ||
| + | pa.adjusted = pmin(pa*length(pa),rep(1,length(pa))) | ||
| + | |||
| + | # there are less conservative multiple testing correction methods, e.g. Holm's method | ||
| + | # but in this case we get almost the same results | ||
| + | pa.adjusted2 = p.adjust(pa, method ="holm") | ||
| + | </pre> | ||
| + | * Other frequently used correction is [https://en.wikipedia.org/wiki/False_discovery_rate false discovery rate (FDR)], which is less strict and controls the overall proportion of false positives among results | ||
| + | =HW06= | ||
| + | [[#L06|Lecture 6]] | ||
| + | |||
| + | * Do either tasks A,B,C (beginners) or B,C,D (more advanced). If you really want, you can do all four for bonus credit. | ||
| + | * In your protocol write used R commands with brief comments on your approach. | ||
| + | * Submit required plots with filenames as specified. | ||
| + | * For each task also include results as required and a short discussion commenting the results/plots you have obtained. Is the value of interest increasing or decreasing with some parameter? Are the results as expected or surprising? | ||
| + | * Outline of protocol is in /tasks/hw06/protocol.txt | ||
| + | |||
| + | ==Task A: sign test== | ||
| + | |||
| + | * Consider a situation in which players played ''n'' games, out of which a fraction of ''q'' were won by ''A'' (example in lecture corresponds to ''q=0.6'' and ''n=10'') | ||
| + | * Compute a table of p-values for ''n=10,20,...,90,100'' and for ''q=0.6, 0.7, 0.8, 0.9'' | ||
| + | * Plot the table using matplot (''n'' is x-axis, one line for each value of ''q'') | ||
| + | * '''Submit''' the plot in <tt>sign.png</tt> | ||
| + | * '''Discuss''' the values you have seen in the plot / table | ||
| + | |||
| + | Outline of the code: | ||
| + | <pre> | ||
| + | # create vector rows with values 10,20,...,100 | ||
| + | rows=(1:10)*10 | ||
| + | # create vector columns with required values of q | ||
| + | columns=c(0.6, 0.7, 0.8, 0.9) | ||
| + | # create empty matrix of pvalues | ||
| + | pvalues = matrix(0,length(rows),length(columns)) | ||
| + | # TODO: fill in matrix pvalues using binom.test | ||
| + | |||
| + | # set names of rows and columns | ||
| + | rownames(pvalues)=rows | ||
| + | colnames(pvalues)=columns | ||
| + | # careful: pvalues[10,] is now 10th row, i.e. value for n=100, | ||
| + | # pvalues["10",] is the first row, i.e. value for n=10 | ||
| + | |||
| + | # check that for n=10 and q=0.6 you get p-value 0.3769531 | ||
| + | pvalues["10","0.6"] | ||
| + | |||
| + | # create x-axis matrix (as in HW05, part D) | ||
| + | x=matrix(rep(rows,length(columns)),nrow=length(rows)) | ||
| + | # matplot command | ||
| + | png("sign.png") | ||
| + | matplot(x,pvalues,type="l",col=c(1:length(columns)),lty=1) | ||
| + | legend("topright",legend=columns,col=c(1:length(columns)),lty=1) | ||
| + | dev.off() | ||
| + | </pre> | ||
| + | |||
| + | ==Task B: Welch's t-test on microarray data== | ||
| + | |||
| + | * Read table with microarray data, transform it to log scale, then work with table ''a'': | ||
| + | <pre> | ||
| + | input=read.table("/tasks/hw06/acids.tsv", header=TRUE, row.names=1) | ||
| + | a = log(input) | ||
| + | </pre> | ||
| + | * Columns 1,2,3 are reference, columns 4,5,6 acetic acid, 7,8,9 benzoate, 10,11,12 propionate, and 13,14,15 sorbate | ||
| + | * Write a function <tt>my.test</tt> which will take as arguments table ''a'' and 2 lists of columns (e.g. 1:3 and 4:6) and will run for each row of the table Welch's t-test of the first set of columns vs the second set. It will return the resulting vector of p-values | ||
| + | * For example by calling <tt>pa <- my.test(a, 1:3, 4:6)</tt> we will compute p-values for differences between reference and acetic acids (computation may take some time) | ||
| + | * The first 5 values of pa should be | ||
| + | <pre> | ||
| + | > pa[1:5] | ||
| + | [1] 0.94898907 0.07179619 0.24797684 0.48204100 0.23177496 | ||
| + | </pre> | ||
| + | * Run the test for all four acids | ||
| + | * '''Report''' how many genes were significant with p-value <= 0.01 for each acid | ||
| + | ** See [[#HW05#Vector_arithmetics|Vector arithmetics in HW05]] | ||
| + | ** You can count TRUE items in a vector of booleans by sum, e.g. <tt>sum(TRUE,FALSE,TRUE)</tt> is 2 | ||
| + | * '''Report''' how many genes are significant for both acetic and benzoate acids? (logical and is written as <tt>&</tt>) | ||
| + | |||
| + | ==Task C: multiple testing correction== | ||
| + | |||
| + | Run the following snippet of code, which works on the vector of p-values <tt>pa</tt> obtained for acetate in task B | ||
| + | <pre> | ||
| + | # adjusts vectors of p-vales from tasks B for using Bonferroni correction | ||
| + | pa.adjusted = p.adjust(pa, method ="bonferroni") | ||
| + | # add this adjusted vector to frame a | ||
| + | a <- cbind(a, pa.adjusted) | ||
| + | # create permutation ordered by pa.adjusted | ||
| + | oa = order(pa.adjusted) | ||
| + | # select from table five rows with the lowest pa.adjusted (using vector oa) | ||
| + | # and display columns containing reference, acetate and adjusted p-value | ||
| + | a[oa[1:5],c(1:6,16)] | ||
| + | </pre> | ||
| + | |||
| + | You should get output like this: | ||
| + | <pre> | ||
| + | ref1 ref2 ref3 acetate1 acetate2 acetate3 pa.adjusted | ||
| + | SUL1 7.581312 7.394985 7.412040 2.1633230 2.05412373 1.9169226 0.004793318 | ||
| + | YMR244W 2.985682 2.975530 3.054001 0.3364722 0.33647224 0.1823216 0.188582576 | ||
| + | DIP5 6.943991 7.147795 7.296955 0.6931472 0.09531018 0.5306283 0.253995075 | ||
| + | YLR460C 5.620401 5.801212 5.502482 3.2425924 3.48431229 3.3843903 0.307639012 | ||
| + | HXT4 2.821379 3.049273 2.772589 7.7893717 8.24446541 8.3041980 0.573813502 | ||
| + | </pre> | ||
| + | |||
| + | Do the same procedure for benzoate p-values and '''report''' the result. '''Comment''' the results for both acids. | ||
| + | |||
| + | ==Task D: volcano plot, test on data generated from null hypothesis== | ||
| + | Draw a [https://en.wikipedia.org/wiki/Volcano_plot_(statistics) volcano plot] for the acetate data | ||
| + | * x-axis of this plot is the difference in the mean of reference and mean of acetate. | ||
| + | ** You can compute row means of a matrix by rowMeans. | ||
| + | * y-axis is -log10 of the p-value (use original p-values before multiple testing correction) | ||
| + | * You can quickly see genes which have low p-values (high on y-axis) and also big difference in mean expression between the two conditions (far from 0 on x-axis). You can also see if acetate increases or decreases expression of these genes. | ||
| + | |||
| + | Now create a simulated dataset sharing some features of the real data but observing the null hypothesis that the mean of reference and acetate are the same for each gene | ||
| + | * Compute vector ''m'' of means for columns 1:6 from matrix ''a'' | ||
| + | * Compute vectors ''sr'' and ''sa'' of standard deviations for reference columns and for acetate columns respectively | ||
| + | ** You can compute standard deviation for each row of a matrix by <tt>apply(some.matrix, 1, sd)</tt> | ||
| + | * For each for each i in 1:6398, create three samples from normal distribution with mean ''m[i]'' and standard deviation ''sr[i]'', and three samples with mean m[i] and deviation sa[i] | ||
| + | ** Use function <tt>rnorm</tt> | ||
| + | * On the resulting matrix apply Welches' t-test and draw the volcano plot. | ||
| + | * How many random genes have p-value <=0.01? Is it roughly what we would expect under the null hypothesis? | ||
| + | |||
| + | Draw histogram of p-values from the real data (reference vs acetate) and from random data. '''Describe''' how they differ. Is it what you would expect? | ||
| + | * use function <tt>hist</tt> | ||
| + | |||
| + | '''Submit''' plots <tt>volcano-real.png</tt>, <tt>volcano-random.png</tt>, <tt>hist-real.png</tt>, <tt>hist-random.png</tt> | ||
| + | (real for real expression data and random for generated data) | ||
| + | =L07= | ||
| + | In this lecture we dive into SQLite3 and Python. | ||
| + | |||
| + | == SQLite3 == | ||
| + | |||
| + | SQLite3 is a simple "database" stored in one file. Think of SQLite not as a replacement for Oracle but as a replacement for fopen(). | ||
| + | Documentation: https://www.sqlite.org/docs.html | ||
| + | |||
| + | You can access sqlite database either from command line: | ||
| + | <pre> | ||
| + | usamec@Darth-Labacus-2:~$ sqlite3 db.sqlite3 | ||
| + | SQLite version 3.8.2 2013-12-06 14:53:30 | ||
| + | Enter ".help" for instructions | ||
| + | Enter SQL statements terminated with a ";" | ||
| + | sqlite> CREATE TABLE test(id integer primary key, name text); | ||
| + | sqlite> .schema test | ||
| + | CREATE TABLE test(id integer primary key, name text); | ||
| + | sqlite> .exit | ||
| + | </pre> | ||
| + | |||
| + | Or from python interface: https://docs.python.org/2/library/sqlite3.html. | ||
| + | |||
| + | == Python == | ||
| + | |||
| + | Python is a perfect language for almost anything. Here is a cheatsheet: http://www.cogsci.rpi.edu/~destem/igd/python_cheat_sheet.pdf | ||
| + | |||
| + | == Scraping webpages == | ||
| + | |||
| + | The simplest tool for scraping webpages is urllib2: https://docs.python.org/2/library/urllib2.html | ||
| + | Example usage: | ||
| + | <pre> | ||
| + | import urllib2 | ||
| + | f = urllib2.urlopen('http://www.python.org/') | ||
| + | print f.read() | ||
| + | </pre> | ||
| + | |||
| + | == Parsing webpages == | ||
| + | |||
| + | We use beautifulsoup4 for parsing html (http://www.crummy.com/software/BeautifulSoup/bs4/doc/). | ||
| + | I recommend following examples at the beginning of the documentation and example about CSS selectors: | ||
| + | http://www.crummy.com/software/BeautifulSoup/bs4/doc/#css-selectors | ||
| + | |||
| + | == Parsing dates == | ||
| + | |||
| + | You have two options. Either use datetime.strptime or use dateutil package (https://dateutil.readthedocs.org/en/latest/parser.html). | ||
| + | |||
| + | == Other usefull tips == | ||
| + | |||
| + | * Don't forget to commit to your sqlite3 database (db.commit()). | ||
| + | * CREATE TABLE IF NOT EXISTS can be usefull at the start of your script. | ||
| + | * Inspect element (right click on element) in Chrome can be very helpful. | ||
| + | * Use screen command for long running scripts. | ||
| + | * All packages are installed on vyuka server. If you are planning using your own laptop, you need to install them using pip (preferably using virtualenv). | ||
| + | =HW07= | ||
| + | * Submit by copying requested files to /submit/hw07/username/ | ||
| + | |||
| + | General goal: Scrape comments from several (hundreds) sme.sk users from last month and store them in SQLite3 database. | ||
| + | |||
| + | ==Task A== | ||
| + | |||
| + | Create SQLite3 "database" with appropriate schema for storing comments from SME.sk discussions. | ||
| + | You will probably need tables for users and comments. You don't need to store which comments replies to which one. | ||
| + | |||
| + | Submit two files: | ||
| + | * db.sqlite3 - the database | ||
| + | * schema.txt - brief description of your schema and rationale behind it | ||
| + | |||
| + | ==Task B== | ||
| + | |||
| + | Build a crawler, which crawls comments in sme.sk discussions. | ||
| + | You have two options: | ||
| + | * For fewer points: Script which gets url of the user (http://ekonomika.sme.sk/diskusie/user_profile.php?id_user=157432) and crawls his comments from last month. | ||
| + | * For more points: Scripts which gets one starting url (either user profile or some discussion, your choice) and automatically discovers users and crawls their comments. | ||
| + | |||
| + | This crawler should store comments in SQLite3 database built in previous task. | ||
| + | Submit following: | ||
| + | * db.sqlite3 - the database | ||
| + | * every python script used for crawling | ||
| + | * README (how to start your crawler) | ||
| + | =L08= | ||
| + | |||
| + | In this lecture we will use Flask and simple text processing utilities from ScikitLearn. | ||
| + | |||
| + | ==Flask== | ||
| + | |||
| + | Flask is simple web server for python (http://flask.pocoo.org/docs/0.10/quickstart/#a-minimal-application) | ||
| + | You can find sample flask application at /tasks/hw08/simple_flask. | ||
| + | Before running change the port number. | ||
| + | You can then access your app at vyuka.compbio.fmph.uniba.sk:4247 (change port number). | ||
| + | |||
| + | There may be problem with access to strange port numbers due to firewalling rules. There are at least two ways to circumvent this: | ||
| + | * Use X forwarding and run web browser directly from vyuka | ||
| + | local_machine> ssh vyuka.compbio.fmph.uniba.sk -XC | ||
| + | vyuka> chromium-browser | ||
| + | * Create SOCKS proxy to vyuka.compbio.fmph.uniba.sk and set SOCKS proxy at that port on your local machine. Then all web traffic goes through vyuka.compbio.fmph.uniba.sk via ssh tunnel. To create SOCKS proxy server on local machine port 8000 to vyuka.compbio.fmph.uniba.sk: | ||
| + | local_machine> ssh vyuka.compbio.fmph.uniba.sk -D 8000 | ||
| + | (keep ssh session open while working) | ||
| + | |||
| + | Flask uses jinja2 (http://jinja.pocoo.org/docs/dev/templates/) templating language for showing html (you can use strings in python but it is painful). | ||
| + | |||
| + | ==Processing text== | ||
| + | |||
| + | Main tool for processing text is CountVectorizer class from ScikitLearn | ||
| + | (http://scikit--learn.org/stable/modules/generated/sklearn.feature_extraction.text.CountVectorizer.html). | ||
| + | It transforms text into bag of words (for each word we get counts). Example: | ||
| + | |||
| + | <pre> | ||
| + | from sklearn.feature_extraction.text import CountVectorizer | ||
| + | |||
| + | vec = CountVectorizer(strip_accents='unicode') | ||
| + | |||
| + | texts = [ | ||
| + | "Ema ma mamu.", | ||
| + | "Zirafa sa vo vani kupe a hneva sa." | ||
| + | ] | ||
| + | |||
| + | t = vec.fit_transform(texts).todense() | ||
| + | |||
| + | print t | ||
| + | |||
| + | print vec.vocabulary_ | ||
| + | </pre> | ||
| + | |||
| + | ==Useful things== | ||
| + | |||
| + | We are working with numpy arrays here (that's array t in example above) | ||
| + | Numpy arrays has also lots of nice tricks. | ||
| + | First lets create two matrices: | ||
| + | <pre> | ||
| + | >>> import numpy as np | ||
| + | >>> a = np.array([[#1,2,3],[4,5,6]]) | ||
| + | >>> b = np.array([[#7,8],[9,10],[11,12]]) | ||
| + | >>> a | ||
| + | array([[#1, 2, 3], | ||
| + | [4, 5, 6]]) | ||
| + | >>> b | ||
| + | array([[# 7, 8], | ||
| + | [ 9, 10], | ||
| + | [11, 12]]) | ||
| + | </pre> | ||
| + | |||
| + | We can sum this matrices or multiply them by some number: | ||
| + | <pre> | ||
| + | >>> 3 * a | ||
| + | array([[# 3, 6, 9], | ||
| + | [12, 15, 18]]) | ||
| + | >>> a + 3 * a | ||
| + | array([[# 4, 8, 12], | ||
| + | [16, 20, 24]]) | ||
| + | </pre> | ||
| + | |||
| + | We can calculate sum of elements in each matrix, or sum by some axis: | ||
| + | <pre> | ||
| + | >>> np.sum(a) | ||
| + | 21 | ||
| + | >>> np.sum(a, axis=1) | ||
| + | array([ 6, 15]) | ||
| + | >>> np.sum(a, axis=0) | ||
| + | array([5, 7, 9]) | ||
| + | </pre> | ||
| + | |||
| + | There is a lot other usefull functions check https://docs.scipy.org/doc/numpy-dev/user/quickstart.html. | ||
| + | |||
| + | This can help you get top words for each user: | ||
| + | http://docs.scipy.org/doc/numpy/reference/generated/numpy.argsort.html#numpy.argsort | ||
| + | =HW08= | ||
| + | * Submit by copying requested files to /submit/hw08/username/ | ||
| + | |||
| + | General goal: | ||
| + | Build a simple website, which lists all crawled users and for each users has a page with simple statistics for given user. | ||
| + | |||
| + | This lesson requires crawled data from previous lesson, if you don't have one, you can find it at (and thank Baska): | ||
| + | /tasks/hw08/db.sqlite3 | ||
| + | |||
| + | Submit source code (web server and preprocessing scripts) and database files. | ||
| + | |||
| + | ==Task A== | ||
| + | |||
| + | Create a simple flask web application which: | ||
| + | * Has a homepage where is a list of all users (with links to their pages). | ||
| + | * Has a page for each user, which has simple information about user: His nickname, number of posts and hist last 10 posts. | ||
| + | |||
| + | ==Task B== | ||
| + | |||
| + | For each user preprocess and store list of his top 10 words and list of top 10 words typical for him (which he uses much more often than other users, come up with some simple heuristics). | ||
| + | Show this information on his page. | ||
| + | |||
| + | ==Task C== | ||
| + | |||
| + | Preprocess and store list of top three similar users for each user (try to come up with some simple definition of similarity based on text in posts). Again show this information on user page. | ||
| + | |||
| + | Bonus: | ||
| + | Try to use some simple topic modeling (e.g. PCA as in TruncatedSVD from scikit-learn) and use it for finding similar users. | ||
| + | =L09= | ||
| + | [[#HW09]] | ||
| + | |||
| + | In this lesson we make simple javascript visualizations. | ||
| + | |||
| + | Your goal is to take examples from here https://developers.google.com/chart/interactive/docs/ | ||
| + | and tweak them for your purposes. | ||
| + | |||
| + | Tips: | ||
| + | * You can output your data into javascript data structures in Flask template. It is a bad practice, but sufficient for this lesson. (Better way is to load JSON through API). | ||
| + | * Remember that you have to bypass the firewall. | ||
| + | =HW09= | ||
| + | [[#L09]] | ||
| + | |||
| + | * Submit by copying requested files to /submit/hw09/username/ | ||
| + | |||
| + | General goal: | ||
| + | Extend user pages from previous project with simple visualizations. | ||
| + | |||
| + | ==Task A== | ||
| + | |||
| + | Show a calendar, which shows during which days was user active (like this https://developers.google.com/chart/interactive/docs/gallery/calendar#overview). | ||
| + | |||
| + | ==Task B== | ||
| + | |||
| + | Show a histogram of comments length (like this https://developers.google.com/chart/interactive/docs/gallery/histogram#example). | ||
| + | |||
| + | ==Task C== | ||
| + | |||
| + | Try showing a word tree for a user (https://developers.google.com/chart/interactive/docs/gallery/wordtree#overview). Try to normalize the text (lowercase, remove accents). CountVectorizer has method build_analyzer, which returns a function, which does this for you. | ||
| + | =L10= | ||
| + | [[#HW10]] | ||
| + | |||
| + | Hadoop is a distributed filesystem and a [https://en.wikipedia.org/wiki/MapReduce map/reduce framework]. | ||
| + | It allows large data sets to be stored across multiple computers and also efficiently processed through the map reduce paradigm. | ||
| + | |||
| + | == Hadoop setup == | ||
| + | |||
| + | In order to use haddop on the server, you must source a script that sets up all the required environment variables needed to run hadoop: | ||
| + | |||
| + | <pre> | ||
| + | # set up hadoop environment | ||
| + | source ~yoyo/hadoop/hadoop-env.sh | ||
| + | </pre> | ||
| + | |||
| + | After that you should be able to start the <code>hadoop</code> command. | ||
| + | |||
| + | == Distributed filesystem == | ||
| + | |||
| + | In order to process any data it must be stored in hadoop's distributed filesystem (HDFS). | ||
| + | Hadoop exposes the usual filesystem manipulation commands: | ||
| + | |||
| + | <pre> | ||
| + | # hadoop filesystem with usual commands | ||
| + | hadoop fs -ls | ||
| + | hadoop fs -ls / | ||
| + | hadoop fs -mkdir input | ||
| + | hadoop fs -cp /input/example/pg100.txt input/ | ||
| + | hadoop fs -ls input | ||
| + | </pre> | ||
| + | |||
| + | Note that paths beginning with / are absolute, without slash are relative to | ||
| + | your home directory which is <code>/user/USERNAME</code>. | ||
| + | |||
| + | To transfer files between the server and hadoop filesystems you can use the | ||
| + | <code>put</code> and <code>get</code> (you can also use the <code>cat</code> | ||
| + | command to get contents of short text files). | ||
| + | |||
| + | <pre> | ||
| + | hadoop fs -put someLocalFile.txt input/ | ||
| + | hadoop fs -get output/part-00000 . | ||
| + | </pre> | ||
| + | |||
| + | == Map Reduce == | ||
| + | |||
| + | Map/reduce is a programming model / framework that allows processing large ammounts of | ||
| + | (distributed) data in a distributed manner. | ||
| + | |||
| + | The system resembles the <code>map</code> nad <code>reduce</code> functions. | ||
| + | Input data (files) is first processed by the map function into key-value pairs, | ||
| + | these are redistributed so that all pairs with the same key are together and | ||
| + | and then processed by the reduce function to produce the final result. | ||
| + | |||
| + | Multiple "mappers" and "reducers" can be run on different machines (i.e. mappers | ||
| + | can be run directly on the nodes where the data resides and thus no copying of data | ||
| + | needs to be done). | ||
| + | |||
| + | === Simple word count example === | ||
| + | |||
| + | * Given a lot of large files distributed over several servers, produce word statistics: for each word how many times it occurs in the files. | ||
| + | * map: for each input file count the occurrence of each word in this file | ||
| + | * the result is a list of items separated by tab: word number | ||
| + | * hadoop will group together map results from all partial files and will put the same keys together | ||
| + | * reduce will sum together adjacent values with each key | ||
| + | |||
| + | ==== Map part ==== | ||
| + | * read file word by word | ||
| + | * for each word from input print this word, tab and 1 (hadoop will group the same words together anyway) | ||
| + | <pre> | ||
| + | echo "abc abc def abc " | ~yoyo/example/map.awk | ||
| + | </pre> | ||
| + | |||
| + | ==== Redistribute / shuffle ==== | ||
| + | Hadoop wil collect together the same keys, i.e. the result is something like this: | ||
| + | <pre> | ||
| + | echo "abc abc def abc " | ~yoyo/example/map.awk | sort | ||
| + | </pre> | ||
| + | |||
| + | ==== Reduce part ==== | ||
| + | Reducer then does something similar to <code>niq -c</code>, i.e. counts adajcent equal words. Here implemented in bash | ||
| + | <pre> | ||
| + | echo "abc abc def abc " | ~yoyo/example/map.awk | sort | ~yoyo/example/reduce.sh | ||
| + | </pre> | ||
| + | |||
| + | |||
| + | ==== Running in hadoop ==== | ||
| + | |||
| + | Normallly Hadoop Map-Reduce transformations are written in Java. However the default installation of hadoop | ||
| + | contains a map-reduce class ([https://hadoop.apache.org/docs/r1.2.1/streaming.html hadoop streaming]) that cat execute | ||
| + | any process and write input to / read output from it. | ||
| + | |||
| + | Another technical problem with this solution is that the executed programs need to be present on all nodes where | ||
| + | the mappers / reducers will be run. They either can be installed on every node or present in the haddop filesystem. | ||
| + | The streaming class has an option (<code>-file</code>) to automatically upload any progra / data files along with the job. | ||
| + | |||
| + | |||
| + | <pre> | ||
| + | # the streaming library | ||
| + | JAR=${HADOOP_PREFIX}/share/hadoop/tools/lib/hadoop-streaming-2.7.3.jar | ||
| + | BINKS="" | ||
| + | |||
| + | # use files in the /input/example directory for input | ||
| + | # place output in the out directory | ||
| + | # use map.awk and reduce.sh as mapper / reducer, upload them to haddop fs along the job | ||
| + | |||
| + | hadoop jar $JAR $BINKS -mapper map.awk -reducer reduce.sh -input /input/example/ -output out -file map.awk -file reduce.sh | ||
| + | </pre> | ||
| + | =HW10= | ||
| + | [[#L10]] | ||
| + | * Submit the protocol, scripts and required output files. | ||
| + | * Before running the <code>hadoop</code> command, source a script the necessary settings / paths: | ||
| + | <pre> | ||
| + | source ~yoyo/hadoop/hadoop-env.sh | ||
| + | </pre> | ||
| + | |||
| + | * The directory <code>/input/portage/logs</code> contains large ammounts of log files from a gentoo instalation. | ||
| + | * Installation of files is recorded in the logs as follows: | ||
| + | <pre> | ||
| + | --- /usr/bin/ | ||
| + | >>> /usr/bin/make -> gmake | ||
| + | >>> /usr/bin/gmake | ||
| + | </pre> | ||
| + | |||
| + | The first line represents a directory, the second records a symlink being created (we want to include these in the list of files), | ||
| + | the third line represents a regular file being installed. | ||
| + | |||
| + | ==Task A== | ||
| + | |||
| + | * Find out names of all the files that were ever installed and put them to a file <code>installed_all.txt</code>. | ||
| + | |||
| + | ==Task B== | ||
| + | |||
| + | * Find out 10 most rewriten / reinstalled files (you can use <code>sort</code> / <code>head</code> to postprocess the results file). Write the results (most reinstalled file first) to the file <code>top_reinstalled.txt</code>. | ||
| + | |||
| + | == Task C== | ||
| + | |||
| + | * For each package find the number of errors that ever happened during its installation. Write the results to file <code>package_errors.txt</code> in the form <code>package_name numErrors</code>, sorted with package with most errors on the top. | ||
| + | * Treat any occurrence of the string <code>error:</code> as an error (including the colon, case insensitive) | ||
| + | * Packages are marked at the start of each log file with a line (note the space at the start of the line): | ||
| + | <pre> | ||
| + | * Package: sys-devel/make-4.1-r1 | ||
| + | </pre> | ||
| + | =L11= | ||
| + | [[#HW11]] | ||
| + | |||
| + | ==Biological story: tiny monkeys== | ||
| + | * [https://en.wikipedia.org/wiki/Common_marmoset Common marmoset] (Callithrix jacchus, Kosmáč bielofúzý) weights only about 1/4 kg | ||
| + | * Most primates are much bigger | ||
| + | * Which marmoset genes differ from other primates and are related to the small size? | ||
| + | * Positive selection scan computes of each gene a p-value, whether it evolved on the marmoset lineage faster | ||
| + | ** Exact details, see papers [http://compbio.fmph.uniba.sk/papers/expanded.php?paper=2008002] and [http://compbio.fmph.uniba.sk/papers/expanded.php?paper=2014008] | ||
| + | * The result is a list of p-values, one for each gene | ||
| + | * Which biological functions are enriched among positively selected genes? Are any of those functions possibly related to body size? | ||
| + | |||
| + | ==Gene functions and GO categories== | ||
| + | |||
| + | Use mysql database "marmoset" on the server. | ||
| + | |||
| + | * We can look at the description of a particular gene: | ||
| + | select * from genes where prot='IGF1R'; | ||
| + | +----------------------------+-------+-------------------------------------------------+ | ||
| + | | transcriptid | prot | description | | ||
| + | +----------------------------+-------+-------------------------------------------------+ | ||
| + | | knownGene.uc010urq.1.1.inc | IGF1R | insulin-like growth factor 1 receptor precursor | | ||
| + | +----------------------------+-------+-------------------------------------------------+ | ||
| + | * In the database, we have stored all the P-values from positive selection tests: | ||
| + | select * from lrtmarmoset where transcriptid='knownGene.uc010urq.1.1.inc'; | ||
| + | +----------------------------+---------------------+ | ||
| + | | transcriptid | pval | | ||
| + | +----------------------------+---------------------+ | ||
| + | | knownGene.uc010urq.1.1.inc | 0.00142731425252827 | | ||
| + | +----------------------------+---------------------+ | ||
| + | * Genes are also assigned functional categories based on automated processes (including sequence similarity to other genes) and manual curation. The corresponding database is maintained by [http://geneontology.org/ Gene Ontology Consortium]. We can use on-line sources to search for these annotations, e.g. [http://amigo1.geneontology.org/cgi-bin/amigo/go.cgi here]. | ||
| + | * We can also download the whole database and preprocess it into usable form: | ||
| + | select * from genes2gocat,gocatdefs where transcriptid='knownGene.uc010urq.1.1.inc' and genes2gocat.cat=gocatdefs.cat; | ||
| + | (results in 50 categories) | ||
| + | * GO categories have a hierarchical structure - see for example category GO:0005524 ATP binding: | ||
| + | select * from gocatparents,gocatdefs where gocatparents.parent=gocatdefs.cat and gocatparents.cat='GO:0005524'; | ||
| + | +------------+------------+---------+------------+-------------------------------+ | ||
| + | | cat | parent | reltype | cat | def | | ||
| + | +------------+------------+---------+------------+-------------------------------+ | ||
| + | | GO:0005524 | GO:0032559 | isa | GO:0032559 | adenyl ribonucleotide binding | | ||
| + | +------------+------------+---------+------------+-------------------------------+ | ||
| + | ... and continuing further up the hierarchy: | ||
| + | | GO:0032559 | GO:0030554 | isa | GO:0030554 | adenyl nucleotide binding | | ||
| + | | GO:0032559 | GO:0032555 | isa | GO:0032555 | purine ribonucleotide binding | | ||
| + | | GO:0030554 | GO:0001883 | isa | GO:0001883 | purine nucleoside binding | | ||
| + | | GO:0030554 | GO:0017076 | isa | GO:0017076 | purine nucleotide binding | | ||
| + | | GO:0032555 | GO:0017076 | isa | GO:0017076 | purine nucleotide binding | | ||
| + | | GO:0032555 | GO:0032553 | isa | GO:0032553 | ribonucleotide binding | | ||
| + | | GO:0001883 | GO:0001882 | isa | GO:0001882 | nucleoside binding | | ||
| + | | GO:0017076 | GO:0000166 | isa | GO:0000166 | nucleotide binding | | ||
| + | | GO:0032553 | GO:0000166 | isa | GO:0000166 | nucleotide binding | | ||
| + | | GO:0001882 | GO:0005488 | isa | GO:0005488 | binding | | ||
| + | | GO:0000166 | GO:0005488 | isa | GO:0005488 | binding | | ||
| + | | GO:0005488 | GO:0003674 | isa | GO:0003674 | molecular_function | | ||
| + | * What else can be under GO:0032559 adenyl ribonucleotide binding? | ||
| + | select * from gocatparents,gocatdefs where gocatparents.cat=gocatdefs.cat and gocatparents.parent='GO:0032559'; | ||
| + | +------------+------------+---------+------------+-------------+ | ||
| + | | cat | parent | reltype | cat | def | | ||
| + | +------------+------------+---------+------------+-------------+ | ||
| + | | GO:0005524 | GO:0032559 | isa | GO:0005524 | ATP binding | | ||
| + | | GO:0016208 | GO:0032559 | isa | GO:0016208 | AMP binding | | ||
| + | | GO:0043531 | GO:0032559 | isa | GO:0043531 | ADP binding | | ||
| + | +------------+------------+---------+------------+-------------+ | ||
| + | |||
| + | ==Mann–Whitney U test== | ||
| + | * also known as Wilcoxon rank-sum test | ||
| + | * In [[#L06|Lecture 6]], we have used Welch's t-test to test if one set of expression measurements for a gene are significantly different from the second set | ||
| + | * This test assumes that both sets come from normal (Gaussian) distributions with unknown parameters | ||
| + | * [https://en.wikipedia.org/wiki/Mann%E2%80%93Whitney_U_test Mann-Whitney U test] is called non-parametric, because it does not make this assumption | ||
| + | * The null hypothesis is that two sets of measurements were generated by the same unknown probability distribution | ||
| + | * Alternative hypothesis: for X from the first distribution and Y from the second P(X>Y) is not equal P(Y>X) | ||
| + | * We will use a one-side version of the alternative hypothesis: P(X>Y) > P(Y>X) | ||
| + | * Compute test statistics U: | ||
| + | ** compare all pairs X, Y (X from first set, Y from second set) | ||
| + | ** if X>Y, add 1 to U | ||
| + | ** if X==Y, add 0.5 | ||
| + | * For large sets, U is approximately normally distributed under the null hypothesis | ||
| + | |||
| + | How to use in R: | ||
| + | <pre> | ||
| + | # generate 20 samples from exponential distrib. with mean 1 | ||
| + | x = rexp(20, 1) | ||
| + | # generate 30 samples from exponential distrib. with mean 1/2 | ||
| + | y = rexp(30, 2) | ||
| + | |||
| + | # test if values of x greater than y | ||
| + | wilcox.test(x,y,alternative="greater") | ||
| + | # W = 441, p-value = 0.002336 | ||
| + | # alternative hypothesis: true location shift is greater than 0 | ||
| + | # W is the U statistics above | ||
| + | |||
| + | # now generate y twice from the same distrib. as x | ||
| + | y = rexp(30, 1) | ||
| + | wilcox.test(x,y,alternative="greater") | ||
| + | # W = 364, p-value = 0.1053 | ||
| + | # relatively small p-value (by chance) | ||
| + | |||
| + | y = rexp(30, 1) | ||
| + | wilcox.test(x,y,alternative="greater") | ||
| + | # W = 301, p-value = 0.4961 | ||
| + | # now much greater p-value | ||
| + | </pre> | ||
| + | |||
| + | Another form of the test, potentially useful for HW: | ||
| + | * have a vector of values x, binary vector b indicating two classes: 0 and 1 | ||
| + | * test if values marked by 0 are greater than values marked by 1 | ||
| + | <pre> | ||
| + | # generate 10 with mean 1, 30 with mean 1/2, 10 with mean 1 | ||
| + | x = c(rexp(10,1),rexp(30,2),rexp(10,1)) | ||
| + | # classes 10x0, 20x1, 10x0 | ||
| + | b = c(rep(0,10),rep(1,30),rep(0,10)) | ||
| + | wilcox.test(x~b,alternative="greater") | ||
| + | |||
| + | # the same test by distributing into subvectors x0 and x1 for classes 0 and 1 | ||
| + | x0 = x[b==0] | ||
| + | x1 = x[b==1] | ||
| + | wilcox.test(x0,x1,alternative="greater") | ||
| + | # should be the same as above | ||
| + | </pre> | ||
| + | =HW11= | ||
| + | [[#L11|Lecture 11]] | ||
| + | |||
| + | * In this task, you can use a combination of any scripting languages (e.g. Perl, Python, R) but also SQL, command-line tools etc. | ||
| + | * Input is in a database | ||
| + | * Submit required text files (optionally also files with figures in bonus part E) | ||
| + | * Also submit any scripts you have written for this HW | ||
| + | * In the protocol, include shell commands you have run | ||
| + | * Outline of protocol is in /tasks/hw11/protocol.txt | ||
| + | |||
| + | ==Available data== | ||
| + | |||
| + | * All data necessary for this task is available in the mysql database 'marmoset' on the server | ||
| + | * You will find password in /tasks/hw11/readme.txt | ||
| + | * You have read-only access to the 'marmoset' database | ||
| + | * For creating temporary tables, etc., you can use database 'temp_youruserid' (e.g. 'temp_mrkvicka54'), where you are allowed to create new tables and store data | ||
| + | * You can address tables in mysql even between databases: to start client with your writeable database as default location, use: | ||
| + | :: <tt>mysql -p temp_mrkvicka54</tt> | ||
| + | * You can then access data in the table 'genes' in the database 'marmoset' simply by using 'marmoset.genes' | ||
| + | |||
| + | Getting data from database: | ||
| + | * If your want to get data from database to a tab-separated file, write a select query, run with -e, redirect output: | ||
| + | :: <tt>mysql -p marmoset -e 'select transcriptid as id, pval from lrtmarmoset' > pvals.tsv</tt> | ||
| + | |||
| + | ==Task A: Find ancestors of each GO category== | ||
| + | * Compute a table (in your temporary db or in a file) which contains all pairs category and its ancestor | ||
| + | ** In table gocatparents you have pairs category, its parent, so you need a transitive closure over this relation | ||
| + | ** SQL is not very good at this, you can try repeated joins until you find no more ancestors | ||
| + | ** Alternatively, you can simply extract data from the database and process them in a language of your choice | ||
| + | * '''Submit''' file sample-anc.txt which contains the list of all ancestors of GO:0042773, one per line, in sorted order | ||
| + | ** There should be 14 such ancestors, excluding this category itself; the first in sorted order is GO:0006091, the last is GO:0055114 | ||
| + | |||
| + | ==Task B: Gene in/out data for each GO category== | ||
| + | * Again consider category GO:0042773 | ||
| + | * Create a list of all genes that occur in table lrtmarmoset | ||
| + | ** for each such gene list three columns separated by tabs: its transcript id, p-value from lrtmarmoset, and an indicator 0/1 | ||
| + | ** the indicator is 1, if this gene occurs in GO:0042773 or one of its subcategories; 0 otherwise | ||
| + | ** to find, which gene occur directly in GO:0042773, use table genes2gocat, subcategories can be found in your table from part A | ||
| + | ** note that genes2gocat contains more genes, we will consder only genes from lrtmarmoset | ||
| + | * '''Submit''' this file sample-genes.tsv | ||
| + | ** Overall, your table should have 13717 genes, out of which 28 have value 1 in the last column | ||
| + | ** The first lines of this list (when sorted alphabetically) might look as follos: | ||
| + | <pre> | ||
| + | ensGene.ENST00000043410.1.inc 1 0 | ||
| + | ensGene.ENST00000158526.1.inc 0.483315913388483 0 | ||
| + | ... | ||
| + | ensGene.ENST00000456284.1 1 1 | ||
| + | ... | ||
| + | </pre> | ||
| + | * Note that in part C, you will need to run this process for each category in the database, so make it sufficiently automated | ||
| + | |||
| + | ==Task C: Run Man-Whitney U test for each GO category== | ||
| + | * Run Man-Whitney U test for each non-trivial category | ||
| + | ** Non-trivial categories are such that at least one of our genes (from lrtmarmoset) is in the category and at least one of our genes is not in the category | ||
| + | ** You should test, if genes in a particular GO category have smaller p-values in positive selection than genes outside the category | ||
| + | ** List of all categories can be obtained from gocatdefs, but not all of them are non-trivial (there are 12455 non-trivial categories) | ||
| + | * '''Submit''' file test.tsv in which each line contains two tab separated values: | ||
| + | ** GO category id | ||
| + | ** p-value from the test | ||
| + | * For partial points test at least the category GO:0042773 from parts A and B | ||
| + | |||
| + | ==Task D: Report significant categories== | ||
| + | * '''Submit''' file report.tsv with 20 most significant GO categories (lowest p-values) | ||
| + | ** For each category list its ID, p-value and description | ||
| + | ** Order them from the most significant | ||
| + | ** Descriptions are in table gocatdefs | ||
| + | |||
| + | * To your protocol, write any '''observations''' you can make | ||
| + | ** Do any reported categories seem interesting to you? | ||
| + | ** Are any reported categories likely related to each other based on their descriptions? | ||
| + | |||
| + | ==Task E (bonus): cluster significant categories== | ||
| + | |||
| + | * Some categories in task D appear similar according to their name | ||
| + | * Try creating k-means or hierarchical clustering of categories | ||
| + | * Represent each category as a binary vector in which for each gene you have one bit indicating if it is in the category | ||
| + | * Thus categories with the same set of genes will have identical vectors | ||
| + | * Try to report results in an appropriate form (table, text, figure), discuss them in the protocol | ||
| + | |||
| + | ==Note== | ||
| + | * In part C, we have done many statistical tests, resulting P-values should be corrected by multiple testing correction from [[#L06|Lecture 6]] | ||
| + | ** This is not required in this homework, but should be done in a real study | ||
Latest revision as of 20:56, 19 February 2018
Website for 2017/17
- 2017-02-21 (TV) Perl, part 1 (basics, input processing): Lecture 1, Homework 1
- 2017-02-28 (TV) Perl, part 2 (external commands, files, subroutines): Lecture 2, Homework 2
- 2017-03-07 (BB) Command-line tools, Perl one-liners: Lecture 3, Homework 3
- 2017-03-14 (BB) Job scheduling and make Lecture 4, Homework 4
- 2017-03-21 (BB) R, part 1 Lecture 5, Homework 5
- 2017-03-28 (BB) R, part 2 Lecture 6, Homework 6
- 2017-04-04 (VB) Python, web crawling, HTML parsing, sqlite3 Lecture 7, Homework 7
- 2017-04-11 Python and SQL for beginners (work on HW07), consultations on project topics
- 2017-04-18 no lecture (Easter)
- 2017-04-25 (VB) Text data processing, flask Lecture 8, Homework 8
- 2017-05-02 (VB) Data visualization in JavaScript Lecture 9, Homework 9
- 2017-05-09 (JS) Hadoop Lecture 10, Homework 10
- 2017-05-16 (BB) More databases, scripting language of your choice Lecture 11, Homework 11
- 2017-05-23 work on projects
Contents
- 1 Kontakt
- 2 Úvod
- 3 Pravidlá
- 4 L01
- 5 Lecture 1: Perl, part 1
- 5.1 Why Perl
- 5.2 Sources of Perl-related information
- 5.3 Hello world
- 5.4 The first input file for today: sequence repeats
- 5.5 A sample Perl program
- 5.6 Variables, types
- 5.7 Strings, regular expressions
- 5.8 Conditionals, loops
- 5.9 Input, output
- 5.10 The second input file for today: DNA sequencing reads (fastq)
- 6 HW01
- 7 L02
- 8 HW02
- 9 L03
- 9.1 Efficient use of command line
- 9.2 Redirecting and pipes
- 9.3 Text file manipulation
- 9.3.1 Commands echo and cat (creating and printing files)
- 9.3.2 Commands head and tail (looking at start and end of files)
- 9.3.3 Commands wc, ls -lh, od (exploring file stats and details)
- 9.3.4 Command grep (getting lines in files or stdin matching a regular expression)
- 9.3.5 Commands sort, uniq
- 9.3.6 Commands diff, comm (comparing files)
- 9.3.7 Commands cut, paste, join (working with columns)
- 9.3.8 Commands split, csplit (splitting files to parts)
- 9.4 Programs sed and awk
- 9.5 Perl one-liners
- 10 HW03
- 11 L04
- 12 HW04
- 13 L05
- 14 HW05
- 15 L06
- 16 HW06
- 17 L07
- 18 HW07
- 19 L08
- 20 HW08
- 21 L09
- 22 HW09
- 23 L10
- 24 HW10
- 25 L11
- 26 HW11
Kontakt
Vyučujúci
- doc. Mgr. Broňa Brejová, PhD. miestnosť M-163
- Mgr. Tomáš Vinař, PhD., miestnosť M-163
- Mgr. Vladimír Boža, miestnosť M-25
- RNDr. Jozef Šiška, PhD., miestnosť I-7
- Konzultácie po dohode emailom
Rozvrh
- Utorok 14:00-16:20 M-217
Úvod
Cieľová skupina
Tento predmet je určený pre študentov 2. ročníka bakalárskeho študijného programu Bioinformatika a pre študentov bakalárskeho a magisterského študijného programu Informatika, obzvlášť ak plánujú na magisterskom štúdiu absolvovať štátnicové zameranie Bioinformatika a strojové učenie. Radi privítame aj študentov iných zameraní a študijných programov, pokiaľ majú požadované (neformálne) prerekvizity.
Predpokladáme, že študenti na tomto predmete už vedia programovať v niektorom programovacom jazyku a neboja sa učiť podľa potreby nové jazyky. Takisto predpokladáme základnú znalosť práce v Linuxe vrátane spúšťania príkazov na príkazovom riadku (mali by ste poznať aspoň základné príkazy na prácu so súbormi a adresármi ako cd, mkdir, cp, mv, rm, chmod a pod.). Hoci väčšina technológií preberaných na tomto predmete sa dá použiť na spracovanie dát z mnohých oblastí, budeme ich často ilustrovať na príkladoch z oblasti bioinformatiky. Pokúsime sa vysvetliť potrebné pojmy, ale bolo by dobre, ak by ste sa orientovali v základných pojmoch molekulárnej biológie, ako sú DNA, RNA, proteín, gén, genóm, evolúcia, fylogenetický strom a pod. Študentom zamerania Bioinformatika a strojové učenie odporúčame absolvovať najskôr Metódy v bioinformatike, až potom tento predmet.
Ak sa chcete doučiť základy používania príkazového riadku, skúste napr. tento tutoriál: http://korflab.ucdavis.edu/bootcamp.html
Cieľ predmetu
Počas štúdia sa naučíte mnohé zaujímave algoritmy, modely a metódy, ktoré sa dajú použiť na spracovanie dát v bioinformatike alebo iných oblastiach. Ak však počas štúdia alebo aj neskôr v zamestnaní budete chcieť tieto metódy použiť na reálne dáta, zistíte, že väčšinou treba vynaložiť značné úsilie na samotné získanie dát, ich predspracovanie do vhodného tvaru, testovanie a porovnávanie rôznych metód alebo ich nastavení a získavanie finálnych výsledkov v tvare prehľadných tabuliek a grafov. Často je potrebné tieto činnosti veľakrát opakovať pre rôzne vstupy, rôzne nastavenia a podobne. Obzvlášť v bioinformatike je možné si nájsť zamestnanie, kde vašou hlavnou náplňou bude spracovanie dát s použitím už hotových nástrojov, prípadne doplnených menšími vlastnými programami. Na tomto predmete si ukážeme niektoré programovacie jazyky, postupy a technológie vhodné na tieto činnosti. Veľa z nich je použiteľných na dáta z rôznych oblastí, ale budeme sa venovať aj špecificky bioinformatickým nástrojom.
Základné princípy
Odporúčame nasledujúci článok s dobrými radami k výpočtovým experimentom:
- Noble WS. A quick guide to organizing computational biology projects. PLoS Comput Biol. 2009 Jul 31;5(7):e1000424.
Niektoré dôležité zásady:
- Citát z článku Noble 2009: "Everything you do, you will probably have to do over again."
- Dobre zdokumentujte všetky kroky experimentu (čo ste robili, prečo ste to robili, čo vám vyšlo)
- Ani vy sami si o pár mesiacov tieto detaily nebudete pamätať
- Snažte sa udržiavať logickú štruktúru adresárov a súborov
- Ak však máte veľa experimentov, môže byť dostačujúce označiť ich dátumom, nevymýšľať stále nové dlhé mená
- Snažte sa vyhýbať manuálnym úpravám medzivýsledkov, ktoré znemožňujú jednoduché zopakovanie experimentu
- Snažte sa detegovať chyby v dátach
- Skripty by mali skončiť s chybovou hláškou, keď niečo nejde ako by malo
- V skriptoch čo najviac kontrolujte, že vstupné dáta zodpovedajú vašim predstavám (správny formát, rozumný rozsah hodnôt atď.)
- Ak v skripte voláte iný program, kontrolujte jeho exit code
- Tiež čo najčastejšie kontrolujte medzivýsledky výpočtu (ručným prezeraním, výpočtom rôznych štatistík a pod.), aby ste odhalili prípadné chyby v dátach alebo vo vašom kóde
Pravidlá
Známkovanie
- Domáce úlohy: 55%
- Návrh projektu: 5%
- Projekt: 40%
Stupnica:
- A: 90 a viac, B:80...89, C: 70...79, D: 60...69, E: 50...59, FX: menej ako 50%
Formát predmetu
- Každý týždeň 3 vyučovacie hodiny, z toho cca prvá je prednáška a na ďalšie dve cvičenia. Na cvičeniach samostatne riešite príklady, ktoré doma dokončíte ako domácu úlohu.
- Cez skúškové obdobie budete odovzdávať projekt. Po odovzdaní projektov sa bude konať ešte diskusia o projekte s vyučujúcimi, ktorá môže ovplyvniť vaše body z projektu.
- Budete mať konto na Linuxovom serveri určenom pre tento predmet. Toto konto používajte len na účely tohto predmetu a snažte sa server príliš svojou aktivitou nepreťažiť, aby slúžil všetkým študentom. Akékoľvek pokusy úmyselne narušiť chod servera budú považované za vážne porušenie pravidiel predmetu.
Domáce úlohy
- Termín DÚ týkajúcej sa aktuálnej prednášky je vždy do 9:00 v deň nasledujúcej prednášky (t.j. väčšinou o necelý týždeň od zadania).
- Domácu úlohu odporúčame začať robiť na cvičení, kde vám môžeme prípadne poradiť. Ak máte otázky neskôr, pýtajte sa vyučujúcich emailom.
- Domácu úlohu môžete robiť na ľubovoľnom počítači, pokiaľ možno pod Linuxom. Odovzdaný kód alebo príkazy by však mali byť spustiteľné na serveri pre tento predmet, nepoužívajte teda špeciálny softvér alebo nastavenia vášho počítača.
- Domáca úloha sa odovzdáva nakopírovaním požadovaných súborov do požadovaného adresára na serveri. Konkrétne požiadavky budú spresnené v zadaní.
- Ak sú mená súborov špecifikované v zadaní, dodržujte ich. Ak ich vymýšľate sami, nazvite ich rozumne. V prípade potreby si spravte aj podadresáre, napr. na jednotlivé príklady.
- Dbajte na prehľadnosť odovzdaného zdrojového kódu (odsadzovanie, rozumné názvy premenných, podľa potreby komentáre)
Protokoly
- Väčšinou bude požadovanou súčasťou úlohy textový dokument nazvaný protokol.
- Protokol môže byť vo formáte .txt alebo .pdf a jeho meno má byť protocol.pdf alebo protocol.txt (nakopírujte ho do odovzdaného adresára)
- Protokol môže byť po slovensky alebo po anglicky.
- V prípade použitia txt formátu a diakritiky ju kódujte v UTF8, ale pre jednoduchosť môžete protokoly písať aj bez diakritiky. Ak je protocol v pdf formáte, mali by sa v ňom dať selektovať texty.
- Vo väčšine úloh dostanete kostru protokolu, dodržujte ju.
Hlavička protokolu, vyhodnotenie
- Na vrchu protokolu uveďte meno, číslo domácej úluhy a vaše vyhodnotenie toho, ako sa vám úlohu podarilo vyriešiť. Vyhodnotenie je prehľadný zoznam všetkých príkladov zo zadania, ktoré ste aspoň začali riešiť a kódov označujúcich ich stupeň dokončenia:
- kód HOTOVO uveďte, ak si myslíte, že tento príklad máte úplne a správne vyriešený
- kód ČASŤ uveďte, ak ste nevyriešili príklad celý a do poznámky za kód stručne uveďte, čo máte hotové a čo nie, prípadne ktorými časťami si nie ste istí.
- kód MOŽNO uveďte, ak príklad máte celý, ale nie ste si istí, či správne. Opäť v poznámke uveďte, čím si nie ste istí.
- kód NIČ uveďte, ak ste príklad ani nezačali riešiť
- Vaše vyhodnotenie je pre nás pomôckou pri bodovaní. Príklady označené HOTOVO budeme kontrolovať námatkovo, k príkladom označeným MOŽNO sa vám pokúsime dať nejakú spätnú väzbu, takisto aj k príkladom označeným ČASŤ, kde v poznámke vyjadríte, že ste mali nejaké problémy.
- Pri vyhodnotení sa pokúste čo najlepšie posúdiť správnosť vašich riešení, pričom kvalita vášho seba-hodnotenia môže vplývať na celkový počet bodov.
Obsah protokolu
- Ak nie je v zadaní určené inak, protokol by mal obsahovať nasledovné údaje:
- Zoznam odovzdaných súborov: o každom súbore uveďte jeho význam a či ste ho vyrobili ručne, získali z externých zdrojov alebo vypočítali nejakým programom. Ak máte väčšie množstvo súborov so systematickým pomenovaním, stačí vysvetliť schému názvov všeobecne. Súbory, ktorých mená sú špecifikované v zadaní, nemusíte v zozname uvádzať.
- Postupnosť všetkých spustených príkazov, prípadne iných krokov, ktorými ste dospeli k získaným výsledkom. Tu uvádzajte príkazy na spracovanie dát a spúšťanie vašich či iných programov. Netreba uvádzať príkazy súvisiace so samotným programovaním (spúšťanie editora, nastavenie práv na spustenie a pod.), s kopírovaním úlohy na server a pod. Uveďte aj stručné komentáre, čo bolo účelom určitého príkazu alebo skupiny príkazov.
- Zoznam zdrojov: (webstránky a pod., ktoré ste pri riešení úlohy použili. Nemusíte uvádzať webstránku predmetu a zdroje odporučené priamo v zadaní.
Celkovo by protokol mal umožniť čitateľovi zorientovať sa vo vašich súboroch a tiež v prípade záujmu vykonať rovnaké výpočty, akými ste dospeli vy k výsledku. Nemusíte písať slohy, stačia zrozumiteľné a prehľadné heslovité poznámky.
Projekty
Cieľom projektu je vyskúšať si naučené zručnosti na konkrétnom projekte spracovania dát. Vašou úlohou je zohnať si dáta, tieto dáta analyzovať niektorými technikami z prednášok, prípadne aj inými technológiami a získané výsledky zobraziť v prehľadných grafoch a tabuľkách. Ideálne je, ak sa vám podarí prísť k zaujímavým alebo užitočným záverom, ale hodnotiť budeme hlavne voľbu vhodného postupu a jeho technickú náročnosť. Rozsah samotného programovania alebo analýzy dát by mal zodpovedať zhruba dvom domácim úlohám, ale celkovo bude projekt náročnejší, lebo na rozdiel od úloh nemáte postup a dáta vopred určené, ale musíte si ich vymyslieť sami a nie vždy sa prvý nápad ukáže ako správny. V projekte môžete využiť aj existujúce nástroje a knižnice, ale pokiaľ možno používajte nástroje spúšťané na príkazovom riadku.
Zhruba v dvoch tretinách semestra budete odovzdávať návrh projektu (formát txt alebo pdf, rozsah 0.5-1 strana). V tomto návrhu uveďte, aké dáta budete spracovávať, ako ich zoženiete, čo je cieľom analýzy a aké technológie plánujete použiť. Ciele a technológie môžete počas práce na projekte mierne pozmeniť podľa okolností, mali by ste však mať počiatočnú predstavu. K návrhu vám dáme spätnú väzbu, pričom v niektorých prípadoch môže byť potrebné tému mierne alebo úplne zmeniť. Za načas odovzdaný vhodný návrh projektu získate 5% z celkovej známky. Návrh odporúčame pred odovzdaním konzultovať s vyučujúcimi.
Cez skúškové obdobie bude určený termín odovzdania projektu. Podobne ako pri domácich úlohách odovzdávajte adresár s požadovanými súbormi:
- Vaše programy a súbory s dátami (veľmi veľké dátové súbory vynechajte)
- Protokol podobne ako pri domácich úlohách
- formát txt alebo pdf, stručné heslovité poznámky
- obsahuje zoznam súborov, podrobný postup pri analýze dát (spustené príkazy), ako aj použité zdroje (dáta, programy, dokumentácia a iná literatúra atď)
- Správu k projektu vo formáte pdf. Na rozdiel od menej formálneho protokolu by správu mal tvoriť súvislý text v odbornom štýle, podobne ako napr. záverečné práce. Môžete písať po slovensky alebo po anglicky, ale pokiaľ možno gramaticky správne. Správa by mala mať tieto časti:
- úvod, v ktorom vysvetlíte ciele projektu, prípadne potrebné poznatky zo skúmanej oblasti a aké dáta ste mali k dispozícii
- stručný popis metód, v ktorom neuvádzajte detailne jednotlivé kroky, skôr prehľad použitého prístupu a jeho zdôvodnenie
- výsledky analýzy (tabuľky, grafy a pod.) a popis týchto výsledkov, prípadne aké závery sa z nich dajú spraviť (nezabudnite vysvetliť, čo znamenajú údaje v tabuľkách, osi grafov a pod.). Okrem finálnych výsledkov analýzy uveďte aj čiastkové výsledky, ktorými ste sa snažili overovať, že pôvodné dáta a jednotlivé časti vášho postupu sa správajú rozumne.
- diskusiu, v ktorej uvediete, ktoré časti projektu boli náročné a na aké problémy ste narazili, kde sa vám naopak podarilo nájsť spôsob, ako problém vyriešiť jednoducho, ktoré časti projektu by ste spätne odporúčali robiť iným než vašim postupom, čo ste sa na projekte naučili a podobne
Projekty môžete robiť aj vo dvojici, vtedy však vyžadujeme rozsiahlejší projekt a každý člen by mal byť primárne zodpovedný za určitú časť projektu, čo uveďte aj v správe. Dvojice odovzdávajú jednu správu, ale po odovzdaní projektu majú stretnutie s vyučujúcimi individuálne.
Ako nájsť tému projektu:
- Môžete spracovať nejaké dáta, ktoré potrebujete do bakalárskej alebo diplomovej práce, prípadne aj dáta, ktoré potrebujte na iný predmet (v tom prípade uveďte v správe, o aký predmet ide a takisto upovedomte aj druhého vyučujúceho, že ste použili spracovanie dát ako projekt pre tento predmet). Obzvlášť pre BIN študentov môže byť tento predmet vhodnou príležitosťou nájsť si tému bakalárskej práce a začať na nej pracovať.
- Môžete skúsiť zopakovať analýzu spravenú v nejakom vedeckom článku a overiť, že dostanete tie isté výsledky. Vhodné je tiež skúsiť analýzu aj mierne obmeniť (spustiť na iné dáta, zmeniť nejaké nastavenia, zostaviť aj iný typ grafu a pod.)
- Môžete skúsiť nájsť niekoho, kto má dáta, ktoré by potreboval spracovať, ale nevie ako na to (môže ísť o biológov, vedcov z iných oblastí, ale aj neziskové organizácie a pod.) V prípade, že takýmto spôsobom kontaktujete tretie osoby, bolo by vhodné pracovať na projekte obzvlášť zodpovedne, aby ste nerobili zlé meno našej fakulte.
- V projekte môžete porovnávať niekoľko programov na tú istú úlohu z hľadiska ich rýchlosti či presnosti výsledkov. Obsahom projektu bude príprava dát, na ktorých budete programy bežať, samotné spúšťanie (vhodne zoskriptované) ako aj vyhodnotenie výsledkov.
- A samozrejme môžete niekde na internete vyhrabať zaujímavé dáta a snažiť sa z nich niečo vydolovať.
Opisovanie
- Máte povolené sa so spolužiakmi a ďalšími osobami rozprávať o domácich úlohách resp. projektoch a stratégiách na ich riešenie. Kód, získané výsledky aj text, ktorý odovzdáte, musí však byť vaša samostatná práca. Je zakázané ukazovať svoj kód alebo texty spolužiakom.
- Pri riešení domácej úlohy a projektu očakávame, že budete využívať internetové zdroje, najmä rôzne manuály a diskusné fóra k preberaným technológiám. Nesnažte sa však nájsť hotové riešenia zadaných úloh. Všetky použité zdroje uveďte v domácich úlohách a projektoch.
- Ak nájdeme prípady opisovania alebo nepovolených pomôcok, všetci zúčastnení študenti získajú za príslušnú domácu úlohu, projekt a pod. nula bodov (t.j. aj tí, ktorí dali spolužiakom odpísať) a prípad ďalej podstúpime na riešenie disciplinárnej komisii fakulty.
Zverejňovanie
Zadania a materiály k predmetu sú voľne prístupné na tejto stránke. Prosím vás ale, aby ste nezverejňovali ani inak nešírili vaše riešenia domácich úloh, ak nie je v zadaní povedané inak. Vaše projekty môžete zverejniť, pokiaľ to nie je v rozpore s vašou dohodou so zadávateľom projektu a poskytovateľom dát.
L01
Lecture 1: Perl, part 1
Why Perl
- From Wikipedia: It has been nicknamed "the Swiss Army chainsaw of scripting languages" because of its flexibility and power, and possibly also because of its "ugliness".
Oficial slogans:
- There's more than one way to do it
- Easy things should be easy and hard things should be possible
Advantages
- Good capabilities for processing text files, regular expressions, running external programs etc.
- Closer to common programming language than shell scripts
- Perl one-liners on the command line can replace many other tools such as sed and awk
- Many existing libraries
Disadvantages
- Quirky syntax
- It is easy to write very unreadable programs (sometimes joking called write-only language)
- Quite slow and uses a lot of memory. If possible do no read entire input to memory, process line by line
Warning: we will use Perl 5, Perl 6 is quite a different language
- In package perl-doc man pages:
- man perlintro introduction to Perl
- man perlfunc list of standard functions in Perle
- perldoc -f split describes function split, similarly other functions
- perldoc -q sort shows answers to commonly asked questions (FAQ)
- man perlretut and man perlre regular expressions
- man perl list of other manual pages about Perl
- The same content on the web http://perldoc.perl.org/
- Various web tutorials e.g. this one
- Books
- Bioperl [3] big library for bioinformatics
- Perl for Windows: http://strawberryperl.com/
Hello world
It is possible to run the code directly from a command line (more later):
perl -e'print "Hello world\n"'
This is equivalen to the following code stored in a file:
#! /usr/bin/perl -w use strict; print "Hello world!\n";
- First line is a path to the interpreter
- Swith -w switches warnings on, e.g. if we manipulate with an undefined value (equivalen to "use warnings;")
- Second line use strict will switch on a more strict syntax checks, e.g. all variables must be defined
- Use of -w and use strict is strongly recommended
- Store the program in a file, e.g. hello.pl
- Make it executable (chmod a+x hello.pl)
- Run it with command ./hello.pl
- Also possible to run as perl hello.pl (e.g. if we don't have the path to the interpreter in the file or the executable bit set)
The first input file for today: sequence repeats
- In genomes some sequences occur in many copies (often not exactly equal, only similar)
- We have downloaded a table containing such sequence repeats on chromosome 2L of the fruitfly Drosophila melanogaster
- It was done as follows: on webpage http://genome.ucsc.edu/ we select drosophila genome, then in main menu select Tools, Table browser, select group: variation and repeats, track: ReapatMasker, region: position chr2L, output format: all fields from the selected table a output file: repeats.txt
- Each line of the file contains data about one repeat in the selected chromosome. The first line contains column names. Columns are tab-separated. Here are the first two lines:
#bin swScore milliDiv milliDel milliIns genoName genoStart genoEnd genoLeft strand repName repClass repFamily repStart repEnd repLeft id 585 778 167 7 20 chr2L 1 154 -23513558 + HETRP_DM Satellite Satellite 1519 1669 -203 1
- The file can be found at our server under filename /tasks/hw01/repeats.txt (17185 lines)
- A small randomly selected subset of the table rows is in file /tasks/hw01/repeats-small.txt (159 lines)
A sample Perl program
For each type of repeat (column 11 of the file when counting from 0) we want to compute the number of repeats of this type
#!/usr/bin/perl -w
use strict;
#associative array (hash), with repeat type as key
my %count;
while(my $line = <STDIN>) { # read every line on input
chomp $line; # delete end of line, if any
if($line =~ /^#/) { # skip commented lines
next; # similar to "continue" in C, move to next iteration
}
# split the input line to columns on every tab, store them in an array
my @columns = split "\t", $line;
# check input - should have at least 17 columns
die "Bad input '$line'" unless @columns >= 17;
my $type = $columns[11];
# increase counter for this type
$count{$type}++;
}
# write out results, types sorted alphabetically
foreach my $type (sort keys %count) {
print $type, " ", $count{$type}, "\n";
}
This program does the same thing as the following one-liner (more on one-liners in two weeks)
perl -F'"\t"' -lane 'next if /^#/; die unless @F>=17; $count{$F[11]}++; END { foreach (sort keys %count) { print "$_ $count{$_}" }}' filename
Variables, types
Scalar variables
- Scalar variables start with $, they can hold undefined value (undef), string, number, reference etc.
- Perl converts automatically between strings and numbers
perl -e'print((1 . "2")+1, "\n")'
13
perl -e'print(("a" . "2")+1, "\n")'
1
perl -we'print(("a" . "2")+1, "\n")'
Argument "a2" isn't numeric in addition (+) at -e line 1.
1
- If we switch on strict parsing, each variable needs to be defined by my, several variables created and initialized as follows: my ($a,$b) = (0,1);
- Usual set of C-style operators, power is **, string concatenation .
- Numbers compared by <, <=, ==, != etc., strings by lt, le, eq, ne, gt, ge,
- Comparison operator $a cmp $b for strings, $a <=> $b for numbers: returns -1 if $a<$b, 0 if they are equal, +1 if $a>$b
Arrays
- Names start with @, e.g. @a
- Access to element 0 in array: $a[0]
- Starts with $, because the expression as a whole is a scalar value
- Length of array scalar(@a). In scalar context, @a is the same thing.
- e.g. for(my $i=0; $i<@a; $i++) { ... }
- If using non-existent indexes, they will be created, initialized to undef (++, += treat undef as 0)
- Stack/vector using functions push and pop: push @a, (1,2,3); $x = pop @a;
- Analogicaly shift and unshift on the left end of the array (slower)
- Sorting
- @a = sort @a; (sorts alphabetically)
- @a = sort {$a <=> $b} @a; (sort numerically)
- { } can contain arbitrary comparison function, $a and $b are the two compared elements
- Array concatenation @c = (@a,@b);
- Swap values of two variables: ($x,$y) = ($y,$x);
- Iterate through values of an array (values can be changed):
perl -e'my @a = (1,2,3); foreach my $val (@a) { $val++; } print join(" ", @a), "\n";'
2 3 4
Associative array (hashes)
- Names start with %, e.g. %b
- Access elemtn with name "X": $b{"X"}
- Write out all elements of associative array %b
foreach my $key (keys %b) {
print $key, " ", $b{$key}, "\n";
}
- Inicialization with constant: %b = ("key1"=>"value1","key2"=>"value2")
- instead of => you can also use ,
- test for existence of a key: if(exists $a{"x"}) {...}
- (other methods will create the queried key with undef value)
Multidimensional arrays, fun with pointers
- Pointer to a variable: \$a, \@a, \%a
- Pointer to an anonymous array: [1,2,3], pointer to an anonymous hash: {"kluc1"=>"hodnota1"}
- Hash of lists:
my %a = ("fruits"=>["apple","banana","orange"], "vegetables"=>["celery","carrot"]}
$x = $a{"fruits"}[1];
push @{$a{"fruits"}}, "kiwi";
my $aref = \%a;
$x = $aref->{"fruits"}[1];
- Module Data::Dumper has function Dumper, which will recursively print complex data structures
Strings, regular expressions
Strings
- Substring: substr($string, $start, $length)
- used also to access individual charaters (use length 1)
- If we omit $length, considers until the end of the string, negative start counted from the end of the stringzaciatok rata od konca,...
- We can also used to replace a substring by something else: substr($str, 0, 1) = "aaa" (replaces the first character by "aaa")
- Length of a string: length($str)
- Splitting a string to parts: split reg_expression, $string, $max_number_of_parts
- if " " instead of regular expression, splits at whitespace
- Connecting parts join($separator, @strings)
- Other useful functions: chomp (removes end of line), index (finds a substring), lc, uc (conversion to lowercase/uppercase), reverse (mirror image), sprintf (C-style formatting)
Regular expressions
- more in [4]
$line =~ s/\s+$//; # remove whitespace at the end of the line
$line =~ s/[0-9]+/X/g; # replace each sequence of numbers with character X
#from the name of the fasta sequence (starting with >) create a string until the first space
#(\S means non-whitespace), the result is stored in $1, as specified by ()
if($line =~ /^\>(\S+)/) { $name = $1; }
perl -le'$X="123 4 567"; $X=~s/[0-9]+/X/g; print $X'
X X X
Conditionals, loops
if(expression) { # [] and () cannot be omitted
commands
} elsif(expression) {
commands
} else {
commands
}
command if expression; # here () not necessary
command unless expression;
die "negative value of x: $x" unless $x>=0;
for(my $i=0; $i<100; $i++) {
print $i, "\n";
}
foreach my $i (0..99) {
print $i, "\n";
}
$x=1;
while(1) {
$x *= 2;
last if $x>=100;
}
- Undefined value, number 0 and strings "" and "0" evaluate as false, but I would recommmend always explicitly using logical values in conditional expressions, e.g. if(defined $x), if($x eq ""), if($x==0) etc.
Input, output
- Reading one line from standard input: $line = <STDIN>
- If no more input data available, returns undef
- See also [5]
- Special idiom while(my $line = <STDIN>) equivalent to while (defined(my $line = <STDIN>))
- iterates through all lines of input
- chomp $line removes "\n", if any from the end of the string
- output to stdout through print or printf
The second input file for today: DNA sequencing reads (fastq)
- DNA sequencing machines can read only short pieces of DNA called reads
- Reads are usually stored in fastq format
- Files can be very large (gigabytes or more), but we will use only a small sample from bacteria Staphylococcus aureus, source [6]
- Each read is on 4 lines:
- line 1: ID of the read and other description, line starts with @
- line 2: DNA sequence, A,C,G,T are bases (nucleotides) of DNA, N means unknown base
- line 3: +
- line 4: quality string, which is the string of the same length as DNA in line 2. Each character represents quality of one base in DNA. If p is the probability that this base is wrong, the quality string will contain character with ASCII value 33+(-10 log p), where log is decimal logarithm. This means that higher ASCII means base of higher quality. Character ! (ASCII 33) means probability 1 of error, character $ (ASCII 36) means 50% error, character + (ASCII 43) is 10% error, character 5 (ASCII 53) is 1% error.
- Note that some sequencing platforms represent qualities differently (see article linked above)
- Our file has all reads of equal length (this is not always the case)
The first 4 reads from file /tasks/hw01/reads-small.fastq
@SRR022868.1845/1 AAATTTAGGAAAAGATGATTTAGCAACATTTAGCCTTAATGAAAGACCAGATTCTGTTGCCATGTTTGAATGCCTTAAACCAGTAGCAGAATCAGTATAAA + IICIIIIIIIIIID%IIII8>I8III1II,II)I+III*II<II,E;-HI>+I0IB99I%%2GI*=?5*&1>'$0;%'+%%+;#'$&'%%$-+*$--*+(% @SRR022868.1846/1 TAGCGTTGTAAAATAAATTTCTAGAATGGAAGTGATGATATTGAAATACACTCAGATCCTGAATGAAAGATTTATTAAAGTTAAGACGAGAGTCTCATTAT + 4CIIIIIIII52I)IIIII0I16IIIII2IIII;IIAII&I6AI+*+&G5&G.@8/6&%&,03:*.$479.91(9--$,*&/3"$#&*'+#&##&$(&+&+
And now start on #HW01
HW01
See Lecture 1
Files
We have 4 input files for this homework. We recommend creating soft links to your working directory as follows:
ln -s /tasks/hw01/repeats-small.txt . # small version of the repeat file ln -s /tasks/hw01/repeats.txt . # full version of the repeat file ln -s /tasks/hw01/reads-small.fastq . # smaller version of the read file ln -s /tasks/hw01/reads.fastq . # bigger version of the read file
We recommend writing your protocol starting from an outline provided in /tasks/hw01/protocol.txt
Submitting
- Directory /submit/hw01/your_username will be created for you
- Copy required files to this directory, including the protocol named protocol.txt or protocol.pdf
- You can modify these files freely until deadline, but after the deadline of the homework, you will lose access rights to this directory
Task A
- Consider the program for counting repeat types in the lecture 1, save it to file repeat-stat.pl
- Extend it to compute the average length of each type of repeat
- Each row of the input table contains the start and end coordinates of the repeat in columns 7 and 6. The length is simply the difference of these two values.
- Output a table with three columns: type of repeat, the number of occurrences, the average length of the repeat.
- Use printf to print these three items right-justified in columns of sufficient width, print the average length to 1 decimal place.
- If you run your script on the small file, the output should look something like this (exact column widths may differ):
./repeat-stat.pl < repeats-small.txt
DNA 5 377.4
LINE 4 410.2
LTR 13 355.4
Low_complexity 22 47.2
RC 8 236.2
Simple_repeat 106 39.0
- Include in your protocol the output when you run your script on the large file: ./repeat-stat.pl < repeats.txt
- Find out on Wikipedia, what acronyms LINE and LTR stand for. Do their names correspond to their lengths? (Write a short answer in the protocol.)
- Submit only your script, repeat-stat.pl
Task B
- Write a script which reformats FASTQ file to FASTA format, call it fastq2fasta.pl
- fastq file should be on standard input, fasta file written to standard output
- FASTA format is a typical format for storing DNA and protein sequences.
- Each sequence consists of several lines of the file. The first line starts with ">" followed by identifier of the sequence and optinally some further description separated by whitespace
- The sequence itself is on the second line, long sequences are split into multiple lines
- In our case, the name of the sequence will be the ID of the read with @ replaced by > and / replaced by _
- you can try to use tr or s operators
- For example, the first two reads of reads.fastq are:
@SRR022868.1845/1 AAATTTAGGAAAAGATGATTTAGCAACATTTAGCCTTAATGAAAGACCAGATTCTGTTGCCATGTTTGAATGCCTTAAACCAGTAGCAGAATCAGTATAAA + IICIIIIIIIIIID%IIII8>I8III1II,II)I+III*II<II,E;-HI>+I0IB99I%%2GI*=?5*&1>'$0;%'+%%+;#'$&'%%$-+*$--*+(% @SRR022868.1846/1 TAGCGTTGTAAAATAAATTTCTAGAATGGAAGTGATGATATTGAAATACACTCAGATCCTGAATGAAAGATTTATTAAAGTTAAGACGAGAGTCTCATTAT + 4CIIIIIIII52I)IIIII0I16IIIII2IIII;IIAII&I6AI+*+&G5&G.@8/6&%&,03:*.$479.91(9--$,*&/3"$#&*'+#&##&$(&+&+
- These should be reformatted as follows:
>SRR022868.1845_1 AAATTTAGGAAAAGATGATTTAGCAACATTTAGCCTTAATGAAAGACCAGATTCTGTTGCCATGTTTGAATGCCTTAAACCAGTAGCAGAATCAGTATAAA >SRR022868.1846_1 TAGCGTTGTAAAATAAATTTCTAGAATGGAAGTGATGATATTGAAATACACTCAGATCCTGAATGAAAGATTTATTAAAGTTAAGACGAGAGTCTCATTAT
- Submit files fastq2fasta.pl and reads-small.fasta
- the latter file is created by running ./fastq2fasta.pl < reads-small.fastq > reads-small.fasta
Task C
- Write a script fastq-quality.pl which for each position in a read computes the average quality
- Standard input has fastq file with multiple reads, possibly of different lengths
- As quality we will use simply ASCII values of characters in the quality string with value 33 substracted, so the quality is -10 log p
- ASCII value can be computed by function ord
- Positions in reads will be numbered from 0
- Since reads can differ in length, some positions are used in more reads, some in fewer
- For each position from 0 up to the highest position used in some read, print three numbers separated by tabs "\t": the position index, the number of times this position was used in reads, average quality at that position with 1 decimal place (you can again use printf)
- The last two lines when you run ./fastq-quality.pl < reads-small.fastq should be
99 86 5.5 100 86 8.6
- Run the following command, which runs your script on the larger file and selects every 10th position. Include the output in your protocol. Do you see any trend in quality values with increasing position? (Include a short comment in protocol.)
./fastq-quality.pl < reads.fastq | perl -lane 'print if $F[0]%10==0'
- Submit only fastq-quality.pl
Task D
- Write script fastq-trim.pl that trims low quality bases from the end of each read and filters out short reads
- This script should read a fastq file from standard input and write trimmed fastq file to standard output
- It should also accept two command-line arguments: character Q and integer L
- We have not covered processing command line arguments, but you can use the code snippet below for this
- Q is the minimum acceptable quality (characters from quality string with ASCII value >= ASCII value of Q are ok)
- L is the minimum acceptable length of a read
- First find the last base in a read which has quality at least Q (if any). All bases after this base will be removed from both sequence and quality string
- If the resulting read has fewer than L bases, it is omitted from the output
You can check your program by the following tests:
- If you run the following two commands, you should get tmp identical with input and thus output of diff should be empty
./fastq-trim.pl '!' 101 < reads-small.fastq > tmp # trim at quality ASCII >=33 and length >=101 diff reads-small.fastq tmp # output should be empty (no differences)
- If you run the following two commands, you should see differences in 4 reads, 2 bases trimmed from each
./fastq-trim.pl '"' 1 < reads-small.fastq > tmp # trim at quality ASCII >=34 and length >=1 diff reads-small.fastq tmp # output should be differences in 4 reads
- If you run the following commands, you should get empty output (no reads meet the criteria):
./fastq-trim.pl d 1 < reads-small.fastq # quality ASCII >=100, length >= 1 ./fastq-trim.pl '!' 102 < reads-small.fastq # quality ASCII >=33 and length >=102
Further runs and submitting
- Run ./fastq-trim.pl '(' 95 < reads-small.fastq > reads-small-filtered.fastq # quality ASCII >= 40
- Submit files fastq-trim.pl and reads-small-filtered.fastq
- If you have done task C, run quality statistics on a trimmed version of the bigger file using command below and include in the protocol the result. Comment in the protocol on differences between statistics on the whole file in part C and D. Are they as you expected?
./fastq-trim.pl 2 50 < reads.fastq | ./fastq-quality.pl | perl -lane 'print if $F[0]%10==0' # quality ASCII >= 50
- Note: you have created tools which can be combined, e.g. you can create quality-trimmed version of the fasta file by first trimming fastq and then converting to fasta (no need to submit these files)
Parsing command-line arguments in this task (they will be stored in variables $Q and $L):
#!/usr/bin/perl -w use strict; my $USAGE = " Usage: $0 Q L < input.fastq > output.fastq Trim from the end of each read bases with ASCII quality value less than the given threshold Q. If the length of the read after trimming is less than L, the read will be omitted from output. L is a non-negative integer, Q is a character "; # check that we have exactly 2 command-line arguments die $USAGE unless @ARGV==2; # copy command-line arguments to variables Q and L my ($Q, $L) = @ARGV; # check that $Q is one character and $L looks like a non-negative integer die $USAGE unless length($Q)==1 && $L=~/^[0-9]+$/;
L02
Motivation: Building Phylogenetic Trees
The task for today will be to build a phylogenetic tree of several species using sequences of several genes.
- A phylogenetic tree is a tree showing evolutionary history of these species. Leaves are target present-day species, internal nodes are their common ancestors.
- Input contains sequences of genes from each species.
- Step 1: Identify ortholog groups. Orthologs are genes from different species that "correspond" to each other. This is done based on sequence similarity and we can use a tool called blast to identify sequence similarities between individual genes. The result of ortholog group identification will be a set of genes, each gene having one sequence from each of the 6 species
chimp_94013 dog_84719 human_15749 macaque_34640 mouse_17461 rat_09232
- Step 2: For each ortholog group, we need to align genes and build a phylogenetic tree for this gene using existing methods. We can do this using tools muscle (for alignment) and phyml (for phylogenetic tree inference).
Unaligned sequences:
>mouse ATGCAGTTCCCGCACCCGGGGCCCGCGGCTGCGCCCGCCGTGGGAGTCCCGCTGTATGCG >rat ATGCAGTTCCCGCACCCGGGGCCCGCGGCTGCGCCCGCCGTCGGAGTCCCGCTGTACGCG >dog ATGCAGTACCACCCCGGGCCGGCGGCGGCGGGCGCCGTGGGGGTGCCGCTGTACGCG >human ATGCAGTACCCGCACCCCGGGCCGGCGGCGGGCGCCGTGGGGGTGCCGCTGTACGCG >chimp ATGCAGTACCCGCACCCCGGGCCGGCGGCGGGCGCCGTGGGGGTGCCGCTGTACGCG >macaque ATGCAGTACCCGCACCCCGGGCGGCGGCCGTGGGGGTGGC
Aligned sequences:
>mouse ATGCAGTTCCCGCACCCGGGGCCCGCGGCTGCGCCCGCCGTGGGAGTCCCGCTGTATGCG >rat ATGCAGTTCCCGCACCCGGGGCCCGCGGCTGCGCCCGCCGTCGGAGTCCCGCTGTACGCG >dog ATGCAGTAC---CACCCCGGGCCGGCGGCGGCGGGCGCCGTGGGGGTGCCGCTGTACGCG >human ATGCAGTACCCGCACCCCGGGC---CGGCGGCGGGCGCCGTGGGGGTGCCGCTGTACGCG >chimp ATGCAGTACCCGCACCCCGGGC---CGGCGGCGGGCGCCGTGGGGGTGCCGCTGTACGCG >macaque ATGCAGTACCCGCACCCCGGGC----------GGCGGCCGTGGGGGTGGC----------
Phylogenetic tree:
(mouse:0.03240286,rat:0.01544553,(dog:0.03632419,(macaque:0.01505050,(human:0.00000001,chimp:0.00000001):0.00627957):0.01396920):0.10645019);
- Step 3: The result of the previous step will be several trees, one for every gene. Ideally, all trees would be identical, showing the real evolutionary history of the six species. But it is not easy to infer the real tree from sequence data, so trees from different genes might differ. Therefore, in the last step, we will build a consensus tree. This can be done by usina interactive tool called phylip.
- Output is a single consensus tree.
Our goal for today is to build a pipeline that automates the whole task.
Opening files
my $in;
open $in, "<", "path/file.txt" or die; # open file for reading
while(my $line = <$in>) {
# process line
}
close $in;
my $out;
open $out, ">", "path/file2.txt" or die; # open file for writing
print $out "Hello world\n";
close $out;
# if we want to append to a file use the following instead:
# open $out, ">>", "cesta/subor2.txt" or die;
# standard files
print STDERR "Hello world\n";
my $line = <STDIN>;
# files as arguments of a function
citaj_subor($in);
citaj_subor(\*STDIN);
Working with files and directories
Working directories or files with automatically generated names are automagically deleted after the program finishes.
use File::Temp qw/tempdir/;
my $dir = tempdir("atoms_XXXXXXX", TMPDIR => 1, CLEANUP => 1 );
print STDERR "Creating temporary directory $dir\n";
open $out,">$dir/myfile.txt" or die;
Copying files
use File::Copy;
copy("file1","file2") or die "Copy failed: $!";
copy("Copy.pm",\*STDOUT);
move("/dev1/fileA","/dev2/fileB");
Other functions for working with file system, e.g. chdir, mkdir, unlink, chmod, ...
Function glob finds files with wildcard characters similarly as on command line (see also opendir, readdir, and File::Find module)
ls *.pl
perl -le'foreach my $f (glob("*.pl")) { print $f; }'
Additional functions for working with file names, paths, etc. in modules File::Spec and File::Basename.
Testing for an existence of a file (more in perldoc -f -X)
if(-r "file.txt") { ... } # is file.txt readable?
if(-d "dir") {.... } # is dir a directory?
Running external programs
my $ret = system("command arguments");
# returns -1 if cannot run command, otherwise pass the return code
my $allfiles = `ls`; # returns the result of a command as a text # cannot test return code
Using pipes
open $in, "ls |";
while(my $line = <$in>) { ... }
open $out, "| wc";
print $out "1234\n";
close $out;'
1 1 5
Command-line arguments
# module for processing options in a standardized way
use Getopt::Std;
# string with usage manual
my $USAGE = "$0 [options] length filename
Options:
-l switch on lucky mode
-o filename write output to filename
";
# all arguments to the command are stored in @ARGV array
# parse options and remove them from @ARGV
my %options;
getopts("lo:", \%options);
# now there should be exactly two arguments in @ARGV
die $USAGE unless @ARGV==2;
# process options
my ($length, $filenamefile) = @ARGV;
# values of options are in the %options array
if(exists $options{'l'}) { print "Lucky mode\n"; }
For long option names, see module Getopt::Long
Defining functions
Defining new functions
sub function_name {
# arguments are stored in @_ array
my ($firstarg, $secondarg) = @_;
# do something
return ($result, $second_result);
}
- Arrays and hashes are usually passed as references: function_name(\@array, \%hash);
- It is advantageous to pass long string as references as well to prevent needless copying: function_name(\$sequence);
- References need to be dereferenced, e.g. substr($$sequence) or $array->[0]
Bioperl
use Bio::Tools::CodonTable;
sub translate
{
my ($seq, $code) = @_;
my $CodonTable = Bio::Tools::CodonTable->new( -id => $code);
my $result = $CodonTable->translate($seq);
return $result;
}
Defining modules
Module with name XXX should be in file XXX.pm.
package shared;
BEGIN {
use Exporter ();
our (@ISA, @EXPORT, @EXPORT_OK);
@ISA = qw(Exporter);
# symbols to export by default
@EXPORT = qw(funkcia1, funkcia2);
}
sub funkcia1 {
...
}
sub funkcia2 {
...
}
#module must return true
1;
Using the module located in the same directory as .pl file:
use FindBin qw($Bin); # $Bin is the directory with the script use lib "$Bin"; # add bin to the library path use shared;
HW02
Biological background and overall approach
The task for today will be to build a phylogenetic tree of several species using sequences of several genes.
- We will use 6 mammals: human, chimp, macaque, mouse, rat and dog
- A phylogenetic tree is a tree showing evolutionary history of these species. Leaves are target present-day species, internal nodes are their common ancestors.
- There are methods to build trees by comparing DNA or protein sequences of several present-day species.
- Our input contains a small selection of gene sequences from each species. In a real project we would start from all genes (cca 20,000 per species) and would do a careful filtration of problematic sequences, but we skip this step here.
- The first step will be to identify which genes from different species "correspond" to each other. More exactly, we are looking for groups of orthologs. To do so, we will use a simple method based on sequence similarity, see details below. Again, in real project, more complex methods might be used.
- The result of ortholog group identification will be a set of genes, each gene having one sequence from each of the 6 species
- Next we will process each gene separately, aligning them and building a phylogenetic tree for this gene using existing methods.
- The result of the previous step will be several trees, one for every gene. Ideally, all trees would be identical, showing the real evolutionary history of the six species. But it is not easy to infer the real tree from sequence data, so trees from different genes might differ. Therefore, in the last step, we will build a consensus tree.
Technical overview
This task can be organized in different ways, but to practice Perl, we will write a single Perl script which takes as an input a set of fasta files, each containing DNA sequences of several genes from a single species and writes on output the resulting consensus tree.
- For most of the steps, we will use existing bioinformatics tools. The script will run these tools and do some additional simple processing.
Temporary directory
- During its run, the script and various tools will generate many files. All these files will be stored in a single temporary directory which can be then easily deleted by the user.
- We will use Perl library File::Temp to create this temporary directory with a unique name so that the script can be run several times simultaneously without clashing filenames.
- The library by default creates the file in /tmp, but instead we will create it in the current directory so that it is not deleted at restart of the computer and so that it can be more easily inspected for any problems
- The library by default deletes the directory when the script finishes but again, to allow inspection by the user, we will leave the directory in place
Restart
- The script will have a command line option for restarting the computation and omitting the time-consuming steps that were already finished
- This is useful in long-running scripts because during development of the script you will want to run it many times as you add more steps. In real usage the computation can also be interrupted for various reasons.
- Our restart capabilities will be quite rudimentary: before running a potentially slow external program, the script will check if the temporary directory contains a non-empty file with the filename matching the expected output of the program. If the file is found, it is assumed to be correct and complete and the external program is not run.
Command line options
- The script should be named build-tree.pl and as command-line arguments, it will get names of the species
- For example, we can run the script as follows: ./build-tree.pl human chimp macaque mouse rat dog
- The first species, in this case human, will be so called reference species (see task A)
- The script needs at least 2 species, otherwise it will write an error message and stop
- For each species X there should be a file X.fa in the current directory, this is also checked by the script
- Restart is specified by command line option -r followed by the name of temporary directory
- Command-line option handling and creation of temporary directory is already implemented in the script you are given.
Input files
- Each input fasta X.fa file contains DNA sequences of several genes from one species X
- Each sequence name on a line starting with > will contain species name, underscore and gene id, e.g. ">human_00008"
- Species name matches name of the file, gene id is unique within the fasta file
- Species names and gene ids do not contain underscore, whitespace or any other special characters
- Sequence of each gene can be split into several lines
Files and submitting
In /tasks/hw02/ you will find the following files:
- 6 fasta files (*.fa)
- skeleton script build-tree.pl
- This script already contains handling of command line options, entire task B, potentially useful functions my_run and my_delete and suggested function headers for individual tasks. Feel free to change any of this.
- outline of protocol protocol.txt
- directory example with files for two different groups of genes
Copy the files to your directory and continue writing the script
Submitting
- Submit the script, protocol protocol.txt or protocol.pdf and temporary directory with all files created in the run of your script on all 6 species with human as reference.
- Since the commands and names of files are specified in the homework, you do not need to write them in the protocol (unless you change them). Therefore it is sufficient if the protocol contains self-assessment and any used information sources other than those linked from this assignment or lectures.
- Submit by copying to /submit/hw02/your_username
Task A: run blast to find similar sequences
- To find orthologs, we use a simple method by first finding local alignments (regions of sequence similarity) between genes from different species
- For finding alignments, we will use tool blast (ubuntu package blast2)
- Example of running blast:
formatdb -p F -i human.fa blastall -p blastn -m 9 -d human.fa -i mouse.fa -e 1e-5
- Example of output file:
# BLASTN 2.2.26 [Sep-21-2011] # Query: mouse_00492 # Database: human.fa # Fields: Query id, Subject id, % identity, alignment length, mismatches, gap openings, q. start, q. end, s. start, s. end, e-value, bit score mouse_22930 human_00008 90.79 1107 102 0 1 1107 1 1107 0.0 1386 mouse_22930 human_34035 80.29 350 69 0 745 1094 706 1055 3e-37 147 mouse_22930 human_34035 79.02 143 30 0 427 569 391 533 8e-07 46.1
(note last column - score)
- For each non-reference species, save the result of blast search in file species.blast in the temporary directory.
Task B: find orthogroups
This part is already implemented in the skeleton file, you don't need to implement or report anything in this task
- Here, we process all the species.blast files to find ortholog groups.
- Matches are symmetric, and there can be multiple matches for the same gene. We are looking for reciprocal best hits: pairs of genes human_A and mouse_B, where mouse_B is the match with the highest score in mouse for human_A and human_A is the best-scoring match in human for mouse_B.
- Some genes in reference species may have no reciprocal best hits in some of the non-reference species.
- Gene in the reference species and all of its reciprocal best hits constitute orthogroup. If the size of an orthogroup is the same as the number of species, we will call it a complete orthogroup
- In file genes.txt in temporary directory list we will list all orthogroups, one per line.
chimp_94013 dog_84719 human_15749 macaque_34640 mouse_17461 rat_09232 chimp_61053 human_18570 macaque_12627 chimp_41364 human_19217 macaque_88256 rat_82436
Task C: create a file for each orthogroup
- For each complete orthogroup, we will create a fasta file with corresponding DNA sequences.
- The file will be located in temporary directory and will be named genename.fa, where genename is the name of the orthogroup gene from reference species.
- The fasta name for each sequence is the name of species, NOT the name of the gene.
>human CTGCGGCTGAGAGAGATGTGTACACTGGGGACGCACTCCGGATCTGCATAGTGACCAAAGAGGGCATCAGGGAGGAAACTGTTTCCTTAAGGAAGGAC >chimp TGCGGCTGAGAGAGATGTGTACACTGGGGACGCACTCCGGATCTGCATAGTGACCAAAGAGGGCATCAGGGAGGAGACTGTTTCCTTAAGGAAGGAC >macaque CTGCGGCTGAGAGAGACGTGTACACTGGGGACGCGCTCCGGATCTGCATAGTGACCAAAGAGGGCATCAGGGAGGAGACTGTTCCCTTAAGGAAGGAC >mouse CAGCCGAGAGGGATGTGTATACTGGAGATGCTCTCAGGATCTGCATCGTGACCAAAGAGGGCATCAGGGAGGAAACTGTTCCCCTGCGGAAAGAC >rat CAGCCGAGAGGGATGTGTACACTGGAGACGCCCTCAGGATCTGCATCGTGACCAAAGAGGGCATCAGGGAGGAGACTGTTCCCCTTCGGAAAGAC >dog GAGGGATGTGTACACTGGGGATGCACTCAGAATCTGCATTGTGACTAAGGAGGGCATCAGGGAGGAGACTGTTCCCCTGAGGAAGGAT
Task D: build tree for each gene
- For each orthogroup, we need to build a phylogenetic tree.
- The result for file genename.fa should be saved in file genename.tree
- Example of how to do this:
# create multiple alignment of the sequences muscle -diags -in genename.fa -out genename.mfa # change format of the multiple alignment readseq -f12 genename.mfa -o=genename.phy -a # run phylogenetic inferrence program phyml -i genename.phy --datatype nt --bootstrap 0 --no_memory_check # rename the result mv genename.phy_phyml_tree.txt genename.tree
- You can view the multiple alignment (*.mfa and *.phy) by using program seaview
- You can view the resulting tree (*.tree) by using program njplot or figtree
Task E: build consensus tree
- Trees built on individual genes can differ from each other.
- Therefore we build a consensus tree: tree that only contains branches present in most gene trees; other branches are collapsed.
- phylip is an "interactive" program for manipulation of trees. Specific command for building consensus trees is
phylip consense
- input file for phylip needs to contain all trees of which consensus should be built, one per line
- text you would type to phylip manually, can be instead passed on the standard input from the script
- store the output tree from phylip in all_trees.consensus in temporary directory and also print it to standard output
L03
Today: using command-line tools and Perl one-liners.
- We will do simple transformations of text files using command-line tools without writing any scripts or longer programs.
- You will record the commands used in your protocol
- We strongly recommend making a log of commands for data processing also outside of this course
- If you have a log of executed commands, you can easily execute them again by copy and paste
- For this reason any comments are best preceded by #
- If you use some sequence of commands often, you can turn it into a script
Most commands have man pages or are described within man bash
Efficient use of command line
Some tips for bash shell:
- use tab key to complete command names, path names etc
- tab completion can be customized [7]
- use up and down keys to walk through history of recently executed commands, then edit and resubmit chosen command
- press ctrl-r to search in the history of executed commands
- at the end of session, history stored in ~/.bash_history
- command history -a appends history to this file right now
- you can then look into the file and copy appropriate commands to your protocol
- various other history tricks, e.g. special variables [8]
- cd - goes to previously visited directory, also see pushd and popd
- ls -lt | head shows 10 most recent files, useful for seeing what you have done last
Instead of bash, you can use more advanced command-line environments, e.g. iPhyton notebook
Redirecting and pipes
# redirect standard output to file command > file # append to file command >> file # redirect standard error command 2>file # redirect file to standard input command < file # do not forget to quote > in other uses grep '>' sequences.fasta # (without quotes rewrites sequences.fasta) # send stdout of command1 to stdin of command2 command1 | command2 # backtick operator executes command, # removes trailing \n from stdout, substitutes to command line # the following commands do the same thing: head -n 2 file head -n `echo 2` file # redirect a string in ' ' to stdin of command head head -n 2 <<< 'line 1 line 2 line 3' # in some commands, file argument can be taken from stdin if denoted as - or stdin # the following compares uncompressed version of file1 with file2 zcat file1.gz | diff - file2
Make piped commands fail properly:
set -o pipefail
If set, the return value of a pipeline is the value of the last (rightmost) command to exit with a non-zero status, or zero if all commands in the pipeline exit successfully. This option is disabled by default, pipe then returns exit status of the rightmost command.
Text file manipulation
Commands echo and cat (creating and printing files)
# print text Hello and end of line to stdout echo "Hello" # interpret backslash combinations \n, \t etc: echo -e "first line\nsecond\tline" # concatenate several files to stdout cat file1 file2
Commands head and tail (looking at start and end of files)
# print 10 first lines of file (or stdin) head file some_command | head # print the first 2 lines head -n 2 file # print the last 5 lines tail -n 5 file # print starting from line 100 (line numbering starts at 1) tail -n +100 file # print lines 81..100 head -n 100 file | tail -n 20
Commands wc, ls -lh, od (exploring file stats and details)
# prints three numbers: number of lines (-l), number of words (-w), number of bytes (-c) wc file # prints size of file in human-readable units (K,M,G,T) ls -lh file # od -a prints file or stdout with named characters # allows checking whitespace and special characters echo "hello world!" | od -a # prints: # 0000000 h e l l o sp w o r l d ! nl # 0000015
Command grep (getting lines in files or stdin matching a regular expression)
# -i ignores case (upper case and lowercase letters are the same) grep -i chromosome file # -c counts the number of matching lines in each file grep -c '^[12][0-9]' file1 file2 # other options (there is more, see the manual): # -v print/count not matching lines (inVert) # -n show also line numbers # -B 2 -A 1 print 2 lines before each match and 1 line after match # -E extended regular expressions (allows e.g. |) # -F no regular expressions, set of fixed strings # -f patterns in a file # (good for selecting e.g. only lines matching one of "good" ids)
- docs: grep
Commands sort, uniq
# some useful options of sort: # -g numeric sort # -k which column(s) to use as key # -r reverse (from largest values) # -s stable # -t fiels separator # sorting first by column 2 numerically, in case of ties use column 1 sort -k 1 file | sort -g -s -k 2,2 # uniq outputs one line from each group of consecutive identical lines # uniq -c adds the size of each group as the first column # the following finds all unique lines and sorts them by frequency from most frequent sort file | uniq -c | sort -gr
Commands diff, comm (comparing files)
diff compares two files, useful for manual checking
- useful options
- -b (ignore whitespace differences)
- -r for comparing directories
- -q for fast checking for identity
- -y show differences side-by-side
comm compares two sorted files
- writes 3 columns:
- 1: lines occurring in the first file
- 2: lines occurring in the second file
- 3: lines occurring in both files
- some columns can be suppressed with -1, -2, -3
- good for finding set intersections and differences
Commands cut, paste, join (working with columns)
- cut selects only some columns from file (perl/awk more flexible)
- paste puts 2 or more files side by side, separated by tabs or other character
- join is a powerful tool for making joins and left-joins as in databases on specified columns in two files
Commands split, csplit (splitting files to parts)
- split splits into fixed-size pieces (size in lines, bytes etc.)
- csplit splits at occurrence of a pattern (e.g. fasta file into individual sequences)
csplit sequences.fa '/^>/' '{*}'
Programs sed and awk
Both programs process text files line by line, allow to do various transformations
# replace text "Chr1" by "Chromosome 1"
sed 's/Chr1/Chromosome 1/'
# prints first two lines, then quits (like head -n 2)
sed 2q
# print first and second column from a file
awk '{print $1, $2}'
# print the line if difference in first and second column > 10
awk '{ if ($2-$1>10) print }'
# print lines matching pattern
awk '/pattern/ { print }'
# count lines
awk 'END { print NR }'
Perl one-liners
Instead of sed and awk, we will cover Perl one-liners
# -e executes commands
perl -e'print 2+3,"\n"'
perl -e'$x = 2+3; print $x, "\n"';
# -n wraps commands in a loop reading lines from stdin or files listed as arguments
# the following is roughly the same as cat:
perl -ne'print'
# how to use:
perl -ne'print' < input > output
perl -ne'print' input1 input2 > output
# lines are stored in a special variable $_
# this variable is default argument of many functions,
# including print, so print is the same as print $_
# simple grep-like commands:
perl -ne 'print if /pattern/'
# simple regular expression modifications
perl -ne 's/Chr(\d+)/Chromosome $1/; print'
# // and s/// are applied by default to $_
# -l removes end of line from each input line and adds "\n" after each print
# the following adds * at the end of each line
perl -lne'print $_, "*"'
# -a splits line into words separated by whitespace and stores them in array @F
# the next example prints difference in numbers stored in the second and first column
# (e.g. interval size if each line coordinates of one interval)
perl -lane'print $F[1]-$F[0]'
# -F allows to set separator used for splitting (regular expression)
# the next example splits at tabs
perl -F '"\t"' -lane'print $F[1]-$F[0]'
# -i replaces each file with a new transformed version (DANGEROUS!)
# the next example removes empty lines from all .txt files in the current directory
perl -lne 'print if length($_)>0' -i *.txt
# the following example replaces sequence of whitespace by exactly one space
# and removes leading and trailing spaces from lines in all .txt files
perl -lane 'print join(" ", @F)' -i *.txt
# END { commands } is run at the very end, after we finish reading input
# the following example computes the sum of interval lengths
perl -lane'$sum += $F[1]-$F[0]; END { print $sum; }'
# similarly BEGIN { command } before we start
# variable $. contains line number. $ARGV name of file or - for stdin
# the following prints filename and line number in front of every line
perl -ne'printf "%s.%d: %s", $ARGV, $., $_' file1 file2
# moving files *.txt to have extension .tsv:
# first print commands
# then execute by hand or replace print with system
# mv -i asks if something is to be rewritten
ls *.txt | perl -lne '$s=$_; $s=~s/\.txt/.tsv/; print("mv -i $_ $s")'
ls *.txt | perl -lne '$s=$_; $s=~s/\.txt/.tsv/; system("mv -i $_ $s")'
HW03
Lecture 1, Lecture 2, Lecture 3
- In this homework, use command-line tools or one-liners in Perl, awk or sed. Do not write any scripts or programs.
- Each task can be split into several stages and intermediate files written to disk, but you can also use pipelines to reduce the number of temporary files.
- Your commands should work also for other input files with the same format (do not try to generalize them too much, but also do not use very specific properties of a particular input, such as the number of lines etc.)
- Include all relevant used commands in your protocol and add a short description of your approach.
- Submit the protocol and required output files.
- Outline of the protocol is in /tasks/hw03/protocol.txt, submit to directory /submit/hw03/yourname
Bonus
- If you are bored, you can try to write solution of Task B using as small number of characters as possible
- In the protocol, include both normal readable form and the condensed form
- Winner with the shortest set of commands gets some bonus points
Task A
- /tasks/hw03/names.txt contains data about several people, one per line.
- Each line consists of given name(s), surname and email separated by spaces.
- Each person can have multiple given names (at least 1), but exactly one surname and one email. Email is always of the form username@uniba.sk.
- The task is to generate file passwords.csv which contains a randomly generated password for each of these users
- The output file has columns separated by commas ','
- The first column contains username extracted from email address, second column surname, third column all given names and fourth column the randomly generated password
- Submit file passwords.csv with the result of your commands.
- In addition to commands that produce the desired file, also run some commands that could help you to detect potential problems in the input.
- Examples of such problems include input lines with fewer than 3 columns, missing @ in email, or fields containing commas.
- For every type of error, run a command which should have a different result for correct and incorrect files, and this output should be short even if the input file is long. In your protocol, explain what output you expect for correct and incorrect files (e.g. you can use grep to find incorrect lines and count their number - in a good file the number would be 0, in a bad file higher than 0)
- Such checks are not necessary in the other tasks, but in a real project it is a good idea to check your input and intermediate files.
Example line from input:
Pavol Országh Hviezdoslav hviezdoslav32@uniba.sk
Example line from output (password will differ):
hviezdoslav32,Hviezdoslav,Pavol Országh,3T3Pu3un
Hints:
- Passwords can be generated using pwgen, we also recommend using perl, wc, paste.
- In Perl, function pop may be useful for manipulating @F and function join for connecting strings with a separator.
Task B
File:
- /tasks/hw03/saccharomyces_cerevisiae.gff contains annotation of the yeast genome
- Downloaded from http://yeastgenome.org/ on 2016-03-09, in particular from [12].
- It was further processed to omit DNA sequences from the end of file.
- The size of the file is 5.6M.
- For easier work, link the file to your directory by ln -s /tasks/hw03/saccharomyces_cerevisiae.gff yeast.gff
- The file is in GFF3 format [13]
- Lines starting with # are comments, other lines contain tab-separated data about one interval of some chromosome in the yeast genome
- Meaning of the first 5 columns:
- column 0 chromosome name
- column 1 source (can be ignored)
- column 2 type of interval
- column 3 start of interval (1-based coordinates)
- column 4 end of interval (1-based coordinates)
- You can assume that these first 5 columns do not contain whitespace
Task:
- For each chromosome, the file contains a line which has in column 2 string chromosome, and the interval is the whole chromosome.
- To file chrosomes.txt, print a tab-separated list of chromosome names and sizes in the same order as in the input
- The last line of chromosomes.txt should list the total size of all chromosomes combined.
- Submit file chromosomes.txt
- Hint: tab is written in Perl as "\t". Command cat may be useful.
- Your output should start and end as follows:
chrI 230218 chrII 813184 ... ... chrXVI 948066 chrmt 85779 total 12157105
Task C
- Continue processing file from task B. Print for each type of interval (column 2), how many times it occurs in the file.
- Sort from the most common to the least common interval types.
- Hint: commands sort and uniq will be useful. Do not forget to skip comments.
- Submit file types.txt with the output formatted as follows:
7058 CDS
6600 mRNA
...
...
1 telomerase_RNA_gene
1 mating_type_region
1 intein_encoding_region
Task D
Overall goal:
- Proteins from several well-studied yeast species were downloaded from database http://www.uniprot.org/ on 2016-03-09
- We have also downloaded proteins from yeast Yarrowia lipolytica. We will pretend that nothing is known about these proteins (as if they were produced by gene finding program in a newly sequenced genome).
- We have run blast of known proteins vs. Y.lip. proteins.
- Now we want to find for each protein in Y.lip. its closest match among all known proteins.
Files:
- /tasks/hw03/known.fa is a fasta file with known proteins from several species
- /tasks/hw03/yarLip.fa is a fasta file with proteins from Y.lip.
- /tasks/hw03/known.blast is the result of running blast of yarLip.fa versus known.fa by these commands:
formatdb -i known.fa blastall -p blastp -d known.fa -i yarLip.fa -m 9 -e 1e-5 > known.blast
- you can link these files to your directory as follows:
ln -s /tasks/hw03/known.fa . ln -s /tasks/hw03/yarLip.fa . ln -s /tasks/hw03/known.blast .
Step 1:
- Get the first (strongest) match for each query from known.blast.
- This can be done by printing the lines that are not comments but follow a comment line starting with #.
- In a perl one-liner, you can create a state variable which will remember if the previous line was a comment and based on that you decide of you print the current line.
- Instead of using perl, you can play with grep. Option -A 1 prints the matching lines as well as one line ofter each match
- Print only the first two columns separated by tab (name of query, name of target), sort the file by second column.
- Submit file best.tsv with the result
- File should start as follows:
Q6CBS2 sp|B5BP46|YP52_SCHPO Q6C8R4 sp|B5BP48|YP54_SCHPO Q6CG80 sp|B5BP48|YP54_SCHPO Q6CH56 sp|B5BP48|YP54_SCHPO
Step 2:
- Submit file known.tsv which contains sequence names extracted from known.fa with leading > removed
- This file should be sorted alphabetically.
- File should start as follows:
sp|A0A023PXA5|YA19A_YEAST Putative uncharacterized protein YAL019W-A OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) GN=YAL019W-A PE=5 SV=1 sp|A0A023PXB0|YA019_YEAST Putative uncharacterized protein YAR019W-A OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) GN=YAR019W-A PE=5 SV=1
Step 3:
- Use command join to join the files best.tsv and known.tsv so that each line of best.tsv is extended with the text describing the corresponding target in known.tsv
- Use option -1 2 to use second column of best.tsv as a key for joining
- The output of join may look as follows:
sp|B5BP46|YP52_SCHPO Q6CBS2 Putative glutathione S-transferase C1183.02 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) GN=SPBC460.02c PE=3 SV=1 sp|B5BP48|YP54_SCHPO Q6C8R4 Putative alpha-ketoglutarate-dependent sulfonate dioxygenase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) GN=SPBC460.04c PE=3 SV=1
- Further reformat the output so that query name goes first (e.g. Q6CBS2), followed by target name (e.g. sp|B5BP46|YP52_SCHPO), followed by the rest of the text, but remove all text after OS=
- Sort by query name
- Submit file best.txt with the result
- The output should start as follows:
B5FVA8 tr|Q5A7D5|Q5A7D5_CANAL Lysophospholipase B5FVB0 sp|O74810|UBC1_SCHPO Ubiquitin-conjugating enzyme E2 1 B5FVB1 sp|O13877|RPAB5_SCHPO DNA-directed RNA polymerases I, II, and III subunit RPABC5
Note:
- not all Y.lip. are necessarily included in your final output (some proteins do not have blast match)
- you can think how to find the list of such proteins, but this is not part of the assignment
- but files best.txt and best.tsv should have the same number of lines
L04
Job Scheduling
- Some computing jobs take a lot of time: hours, days, weeks,...
- We do not want to keep a command-line window open the whole time; therefore we run such jobs in the background
- Simple commands to do it in Linux: batch, at, screen
- These commands run jobs immediately (screen, tmux) or at preset time (at), or when the computer becomes idle (batch)
- Now we will concentrate on Sun Grid Engine, a complex software for managing many jobs from many users on a cluster from multiple computers
- Basic workflow:
- Submit a job (command) to a queue
- The job waits in the queue until resources (memory, CPUs, etc.) become available on some computer
- Then the job runs on the computer
- Output of the job stored in files
- User can monitor the status of the job (waiting, running)
- Complex possibilities for assigning priorities and deadlines to jobs, managing multiple queues etc.
- Ideally all computers in the cluster share the same environment and filesystem
- We have a simple training cluster for this exercise:
- You submit jobs to queue on vyuka
- They will run on computer cpu02 with 8 CPUs
- This cluster is only temporarily available until next Wednesday
Submitting a job (qsub)
- qsub -b y -cwd 'command < input > output 2> error'
- quoting around command allows us to include special characters, such as <, > etc. and not to apply it to qsub command itself
- -b y treats command as binary, usually preferable for both binary programs and scripts
- -cwd executes command in the current directory
- -N name allows to set name of the job
- -l resource=value requests some non-default resources
- for example, we can use -l threads=2 to request 2 threads for parallel programs
- Grid engine will not check if you do not use more CPUs or memory than requested, be considerate (and perhaps occassionally watch your jobs by running top at the computer where they execute)
- qsub will create files for stdout and stderr, e.g. s2.o27 and s2.e27 for the job with name s2 and jobid 27
Monitoring and deleting jobs (qstat, qdel)
- qstat displays jobs of the current user
job-ID prior name user state submit/start at queue slots ja-task-ID
-----------------------------------------------------------------------------------------------------------------
28 0.50000 s3 bbrejova r 03/15/2016 22:12:18 main.q@cpu02.compbio.fmph.unib 1
29 0.00000 s3 bbrejova qw 03/15/2016 22:14:08 1
- qstat -u '*' displays jobs of all users
- finished jobs disappear from the list
- qstat -F threads shows how many threads available
queuename qtype resv/used/tot. load_avg arch states
---------------------------------------------------------------------------------
main.q@cpu02.compbio.fmph.unib BIP 0/2/8 0.03 lx26-amd64
hc:threads=0
28 0.75000 s3 bbrejova r 03/15/2016 22:12:18 1
29 0.25000 s3 bbrejova r 03/15/2016 22:14:18 1
- Command qdel allows you to delete a job (waiting or running)
Interactive work on the cluster (qrsh), screen
- qrsh creates a job which is a normal interactive shell running on the cluster
- in this shell you can then manually run commands
- however, when you close the shell, the job finishes
- therefore it is a good idea to run qrsh within screen
- run screen command, this creates a new shell
- within this shell, run qrsh, then whatever commands
- by pressing Ctrl-a d you "detach" the screen, so that both shells (local and qrsh) continue running but you can close your local window
- later by running screen -r you get back to your shells
Running many small jobs
For example tens of thousands of genes, runs some computation for each gene
- Have a script which iterates through all and runs them sequentially (as in HW02).
- Problems: Does not use parallelism, needs more programming to restart after some interruption
- Submit processing of each gene as a separate job to cluster (submitting done by a script/one-liner).
- Jobs can run in parallel on many different computers
- Problem: Queue gets very long, hard to monitor progress, hard to resubmit only unfinished jobs after some failure.
- Array jobs in qsub (option -t): runs jobs numbered 1,2,3...; number of the job in an environment variable, used by the script to decide which gene to process
- Queue contains only running sub-jobs plus one line for the remaining part of the array job.
- After failure, you can resubmit only unfinished portion of the interval (e.g. start from job 173).
- Next: using make in which you specify how to process each gene and submit a single make command to the queue
- Make can execute multiple tasks in parallel using several threads on the same computer (qsub array jobs can run tasks on multiple computers)
- It will automatically skip tasks which are already finished
Make
- Make is a system for automatically building programs (running compiler, linker etc)
- In particular, we will use GNU make
- Rules for compilation are written in a Makefile
- Rather complex syntax with many features, we will only cover basics
Rules
- The main part of a Makefile are rules specifying how to generate target files from some source files (prerequisites).
- For example the following rule generates target.txt by concatenating source1.txt a source2.txt:
target.txt : source1.txt source2.txt
cat source1.txt source2.txt > target.txt
- The first line describes target and prerequisites, starts in the first column
- The following lines list commands to execute to create the target
- Each line with a command starts with a tab character
- If we now have a directory with this rule in Makefile and files source1.txt and source2.txt, running make target.txt will run the cat command
- However, if target.txt already exists, the command will be run only if one of the prerequisites has more recent modification time than the target
- This allows to restart interrupted computations or rerun necessary parts after modification of some input files
- Makefile automatically chains the rules as necessary:
- if we run make target.txt and some prerequisite does not exist, Makefile checks if it can be created by some other rule and runs that rule first
- In general it first finds all necessary steps and runs them in topological order so that each rules has its prerequisites ready
- Option make -n target will show what commands would be executed to build target (dry run) - good idea before running something potentially dangerous
Pattern rules
- We can specify a general rule for files with a systematic naming scheme. For example, to create a .pdf file from a .tex file, we use pdflatex command:
%.pdf : %.tex
pdflatex $^
- In the first line, % denotes some variable part of the filename, which has to agree in the target and all prerequisites
- In commands, we can use several variables:
- $^ contains name for the prerequisite (source)
- $@ contains the name of the target
- $* contains the string matched by %
Other useful tricks in Makefiles
Variables
- Store some reusable values in variables, then use them several times in the Makefile:
PATH := /projects/trees/bin
target : source
$(PATH)/script < $^ > $@
Wildcards, creating a list of targets from files in the directory
The following Makefile automatically creates .png version of each .eps file simply by running make:
EPS := $(wildcard *.eps)
EPSPNG := $(patsubst %.eps,%.png,$(EPS))
all: $(EPSPNG)
clean:
rm $(EPSPNG)
%.png : %.eps
convert -density 250 $^ $@
- variable EPS contains names of all files matching *.eps
- variable EPSPNG contains desirable names of png files
- it is created by taking filenams in EPS and changing.eps to .png
- all if a "phony target" which is not really created
- its rule has no commands but all png files are prerequisites, so are done first
- the first target in Makefile (in this case all) is default when no other target specified on command-line
- clean is also phony target for deleting generated png files
Useful special built-in target names
Include these lines in your Makefile if desired
.SECONDARY: # prevents deletion of intermediate targets in chained rules .DELETE_ON_ERROR: # delete targets if a rule fails
Parallel make
- running make with option -j 4 will run up to 4 commands in parallel if their dependencies are already finished
- easy parallelization on a single computer
Alternatives to Makefiles
- Bioinformatics often uses "pipelines" - sequences of commands run one after another, e.g. by a script of Makefile
- There are many tools developed for automating computational pipelines, see e.g. this review: Jeremy Leipzig; A review of bioinformatic pipeline frameworks. Brief Bioinform 2016 bbw020.
- For example Snakemake
- Workflows can contain shell commands or Python code
- Big advantage compared to Make: pattern rules my contain multiple variable portions (in make only one % per filename)
- For example, you have several fasta files and several HMMs representing protein families and you wans to run each HMM on each fasta file:
rule HMMER:
input: "{filename}.fasta", "{hmm}.hmm"
output: "{filename}_{hmm}.hmmer"
shell: "hmmsearch --domE 1e-5 --noali --domtblout {output} {input[1]} {input[0]}"
HW04
In this homework, we will return to the example in homework 2, where we took genes from several organisms, found orthogroups of corresponding genes and built a phylogenetic tree for each orthogroup. This was all done in a single big Perl script. In this homework, we will write a similar pipeline using make and execute it remotely using qsub. We will use proteins instead of DNA and we will use a different set of species. Most of the work is already done, only small modifications are necessary.
- Submit by copying requested files to /submit/hw04/username/
- Do not forget to submit protocol, outline of the protocol is in /tasks/hw04/protocol.txt
Task A
- In this task, you will run a long alignment job (>1 hour)
- Copy directory /tasks/hw04/large to your home directory
- ref.fa: all proteins from yeast Yarrowia lipolytica
- other.fa: all proteins from 8 other yeast species
- Makefile: run blast on ref.fa vs other.fa (also formats database other.fa before that)
- run make -n to see what commands will be done (you should see formatdb and blastall + echo for timing), copy the output to the protocol
- run qsub with appropriate options to run make (at least -cwd and -b y)
- then run qstat > queue.txt
- Submit file queue.txt showing your job waiting or running
- When your job finishes, submit also the following two files:
- the last 100 lines from the output file ref.blast under the name ref-end.blast (use tool tail -n 100)
- standard output from the qsub job, which is stored in a file named e.g. make.oX where X is the number of your job. The output shows the time when your job started and finished (this information was written by commands echo in the Makefile)
Task B
- In this task, you will finish a Makefile for splitting blast results into orthogroups and building phylogenetic trees for each group
- This Makefile works with much smaller files and so you can run it many times on vyuka, without qsub
- If it runs too slowly, you can temporarily modify ref.fa to contain only the first 3 sequences, debug your makefile and then again copy the original ref.fa from /tasks/hw04/small to run the final analysis
- Copy directory /tasks/hw04/small to your home directory
- ref.fa: 10 proteins from yeast Yarrowia lipolytica
- other.fa: a selected subset of proteins from 8 other yeast species
- Makefile: longer makefile
The Makefile runs the analysis in four stages. Stages 1,2 and 4 are done, you have to finish stage 3
- If you run make without argument, it will attempt to run all 4 stages, but stage 3 will not run, because it is missing
- Stage 1: run as make ref.brm
- It runs blast as in task A, then splits proteins into orthogroups and creates one directory for each group with file prot.fa containing protein sequences
- Stage 2: run as make alignments
- In each directory with a single gene, it will create an alignment prot.phy and link it under names lg.phy and wag.phy
- Stage 3: run as make trees (needs to be written by you)
- In each directory with a single gene, it should create lg.phy_phyml_tree and wag.phy_phyml_tree
- These corresponds to results of phyml commands run with two different evolutionary models WAG and LG, where LG is the default
- Run phyml by commands of the forms:
- phyml -i INPUT --datatype aa --bootstrap 0 --no_memory_check >LOG
- phyml -i INPUT --model WAG --datatype aa --bootstrap 0 --no_memory_check >LOG
- Change INPUT and LOG in the commands to appropriate filenames using make variables $@, $^, $* etc. Input should come from lg.phy or wag.phy in the directory of a gene and log should be the same as tree name with extension .log added (e.g. lg.phy_phyml_tree.log)
- Also add variables LG_TREES and WAG_TREES listing filenames of all desirable trees and uncomment phony target trees wich uses these variables
- Stage 4: run as make consensus
- Output trees from stage 3 are concatenated for each model separately to files lg/intree wag/intree and then phylip is run to produce consensus trees lg.tree and wag.tree
- This stage also needs variables LG_TREES and WAG_TREES to be defined by you.
- Run your Makefile
- Submit the whole directory small, including Makefile and all gene directories with tree files.
- Look at the two resulting trees using the figtree program, switch on displaying branch labels in the left panel with options. These labels show for each branch of the tree, how many of the input trees support this branch. Write to your protocol, what do you observe: Do the two trees differ? If yes, do they differ in branches supported by many different genes trees, or few? What is the highest and lowest support for a branch in each tree?
Further possibilities
Here are some possibilities for further experiments, in case you are interested (do not submit these):
- You could copy your extended Makefile to directory large and create trees for all orthogroups in the big set
- This would take a long time, so submit it through qsub and only some time after the lecture is over to allow classmates to work on task A
- After ref.brm si done, programs for individual genes can be run in parallel, so you can try running make -j 2 and request 2 threads from qsub
- Phyml also supports other models, for example JTT (see manual), you could try to play with those.
- Command touch FILENAME will change modification time of the given file to current file
- What happens when you run touch on some of the intermediate files in the analysis in task B? Does Makefile always run properly?
L05
Program for today: basics of R (applied to biology examples)
- very short intro as a lecture
- tutorial as HW: read a bit of text, try some commands, extend/modify them as requested
In this course we cover several languages popular for scripting in bioinformatics: Perl, Python, R
- their capabilities overlap, many extensions emulate strengths of one in another
- choose a language based on your preference, level of knowledge, existing code for the task, rest of the team
- quickly learn a new language if needed
- also possibly combine, e.g. preprocess data in Perl or Python, then run statistical analyses in R, automate entire pipeline with bash or make
Introduction
- R is an open-source system for statistical computing and data visualization
- Programming language, command-line interface
- Many built-in functions, additional libraries
- For example http://bioconductor.org/ for bioinformatics
- We will concentrate on useful commands rather than language features
Working in R
- Run command R, type commands in command-line interface
- supports history of commands (arrows, up and down, Ctrl-R) and completing command names with tab key
> 1+2 [1] 3
- Write a script to file, run it from command-line: R --vanilla --slave < file.R
- Use rstudio to open a graphics IDE [14]
- Windows with editor of R scripts, console, variables, plots
- Ctrl-Enter in editor executes current command in console
x=c(1:10) plot(x,x*x)
- ? plot displays help for plot command
Suggested workflow
- work interactively in Rstudio or on command line, try various options
- select useful commands, store in a script
- run script automatically on new data/new versions, potentially as a part of a bigger pipeline
Additional information
- Official tutorial
- Seefeld, Linder: Statistics Using R with Biological Examples (pdf book)
- Patrick Burns: The R Inferno (intricacies of the language)
- Other books
Gene expression data
- Gene expression: DNA->mRNA->protein
- Level of gene expression: Extract mRNA from a cell, measure amounts of mRNA
- Technologies: microarray, RNA-seq
Gene expression data
- Rows: genes
- Columns: experiments (e.g. different conditions or different individuals)
- Each value is expression of a gene, i.e. relative amount of mRNA for this gene in the sample
We will use microarray data for yeast:
- Strassburg, Katrin, et al. "Dynamic transcriptional and metabolic responses in yeast adapting to temperature stress." Omics: a journal of integrative biology 14.3 (2010): 249-259. [15]
- Downloaded from GEO database [16]
- Data already preprocessed: normalization, log2, etc
- We have selected only cold conditions, genes with absolute change at least 1
- Data: 2738 genes, 8 experiments in a time series, yeast moved from normal temperature 28 degrees C to cold conditions 10 degrees C, samples taken after 0min, 15min, 30min, 1h, 2h, 4h, 8h, 24h in cold
HW05
In this homework, try to read text, execute given commands, potentially trying some small modifications.
- Then do tasks A-D, submit required files (3x .png)
- In your protocol, enter commands used in tasks A-D, with explanatory comments in more complicated situations
- In task B also enter required output to protocol
First steps
- Type a command, R writes the answer, e.g.:
> 1+2 [1] 3
- We can store values in variables and use them later on
> # The size of the sequenced portion of cow's genome, in millions of base pairs > Cow_genome_size <- 2290 > Cow_genome_size [1] 2290 > Cow_chromosome_pairs <- 30 > Cow_avg_chrom <- Cow_genome_size / Cow_chromosome_pairs > Cow_avg_chrom [1] 76.33333
Surprises:
- dots are used as parts of id's, e.g. read.table is name of a single function (not method for object read)
- assignment via <- or =
- careful: a<-3 is assignment, a < -3 is comparison
- vectors etc are indexed from 1, not from 0
Vectors, basic plots
- Vector is a sequence of values of the same type (all are numbers or all are strings or all are booleans)
# Vector can be created from a list of numbers by function c
a<-c(1,2,4)
a
# prints [1] 1 2 4
# function c also concatenates vectors
c(a,a)
# prints [1] 1 2 4 1 2 4
# Vector of two strings
b<-c("hello", "world")
# Create a vector of numbers 1..10
x<-1:10
x
# prints [1] 1 2 3 4 5 6 7 8 9 10
Vector arithmetics
- Operations applied to each member of the vector
x<-1:10 # Square each number in vector x x*x # prints [1] 1 4 9 16 25 36 49 64 81 100 # New vector y: logarithm from a number in x squared y<-log(x*x) y # prints [1] 0.000000 1.386294 2.197225 2.772589 3.218876 3.583519 3.891820 4.158883 # [9] 4.394449 4.605170 # Draw graph of function log(x*x) for x=1..10 plot(x,y) # The same graph but use lines instead of dots plot(x,y,type="l") # Addressing elements of a vector: positions start at 1 # Second element of the vector y[2] # prints [1] 1.386294 # Which elements of the vector satisfy certain condition? (vector of logical values) y>3 # prints [1] FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE TRUE # write only those elements from y that satisfy the condition y[y>3] # prints [1] 3.218876 3.583519 3.891820 4.158883 4.394449 4.605170 # we can also write values of x such that values of y satisfy the condition... x[y>3] # prints [1] 5 6 7 8 9 10
- Alternative plotting facilities: ggplot2 library, lattice library
Task A
- Create a plot of the binary logarithm with dots in the graph more densely spaced (from 0.1 to 10 with step 0.1)
- Store it in file log.png and submit this file
- Hints:
- Create x and y by vector arithmetics
- To compute binary logarithm check help ? log
- Before running plot, use command png("log.png") to store the result, afterwards call dev.off() to close the file (in rstudio you can also export plots manually)
Data frames and simple statistics
- Data frame: table similar to spreadsheet, each column is a vector, all are of the same length
- We will use a table with the following columns:
- The size of the sequenced portion of a genome, in millions of base pairs
- Number of chromosome pairs
- GC content
- taxonomic group mammal or fish
- Stored in CSV format, columns separated by tabs.
- Data: Han et al Genome Biology 2008 [17]
Species Size Chrom GC Group Human 2850 23 40.9 mammal Chimpanzee 2750 24 40.7 mammal Macaque 2650 21 40.7 mammal Mouse 2480 20 41.7 mammal ... Tetraodon 187 21 45.9 fish ...
# reading a frame from file
a<-read.table("/tasks/hw05/genomes.csv", header = TRUE, sep = "\t");
# column with name size
a$Size
# Average chromosome length: divide size by the number of chromosomes
a$Size/a$Chrom
# Add average chromosome length as a new column to frame a
a<-cbind(a,AvgChrom=a$Size/a$Chrom)
# scatter plot average chromosome length vs GC content
plot(a$AvgChrom, a$GC)
# compactly display structure of a
# (good for checking that import worked etc)
str(a)
# display mean, median, etc. of each column
summary(a);
# average genome size
mean(a$Size)
# average genome size for mammals
mean(a$Size[a$Group=="mammal"])
# Standard deviation
sd(a$Size)
# Histogram of genome sizes
hist(a$Size)
Task B
- Divide frame a to two frames, one for mammals, one for fish. Hint:
- Try command a[c(1,2,3),]. What is it doing?
- Try command a$Group=="mammal".
- Combine these two commands to get rows for all mammals and store the frame in a new variable, then repeat for fish
- Use a general approach which does not depend on the exact number and ordering of rows in the table.
- Run the command summary separately for mammals and for fish. Which of their characteristics are different?
- Write output and your conclusion to the protocol
Task C
- Draw a graph comparing genome size vs GC content; use different colors for points representing mammals and fish
- Submit the plot in file genomes.png
- To draw the graph, you can use one of the options below, or find yet another way
- Option 1: first draw mammals with one color, then add fish in another color
- Color of points can be changed by: plot(1:10,1:10, col="red")
- After plot command you can add more points to the same graph by command points, which can be used similarly as plot
- Warning: command points does not change the ranges of x and y axes. You have to set these manually so that points from both groups are visible. You can do this using options xlim and ylim, e.g. plot(x,y, col="red", xlim=c(1,100), ylim=c(1,100))
- Option 2: plot both mammals and fish in one plot command, and give it a vector of colors, one for each point
- plot(1:10,1:10,col=c(rep("red",5),rep("blue",5))) will plot the first 5 points red and the last 5 points blue
- Bonus task: add a legend to the plot, showing which color is mammal and which is fish
Expression data and clustering
Data here is bigger, better to use plain R rather than rstudio (limited server CPU/memory)
# Read gene expression data table
a<-read.table("/tasks/hw05/microarray.csv", header = TRUE, sep = "\t", row.names=1)
# Visual check of the first row
a[1,]
# plot starting point vs. situation after 1 hour
plot(a$cold_0min,a$cold_1h)
# to better see density in dense clouds of points, use this plot
smoothScatter(a$cold_15min,a$cold_1h)
# outliers away from diagonal in the plot above are most strongly differentially expressed genes
# these are more easy to see in MA plot:
# x-axis: average expression in the two conditions
# y-axis: difference between values (they are log-scale, so difference 1 means 2-fold)
smoothScatter((a$cold_15min+a$cold_1h)/2,a$cold_15min-a$cold_1h)
Clustering is a wide group of methods that split data points into groups with similar properties
- We will group together genes that have a similar reaction to cold, i.e. their rows in gene expression data matrix have similar values
We will consider two simple clustering methods
- K means clustering splits points (genes) into k clusters, where k is a parameter given by the user. It finds a center of each cluster and tries to minimize the sum of distances from individual points to the center of their cluster. Note that this algorithm is randomized so you will get different clusters each time.
- Hierarchical clustering puts all data points (genes) to a hierarchy so that smallest subtrees of the hierarchy are the most closely related groups of points and these are connected to bigger and more loosely related groups.
# Heatmap: creates hierarchical clustering of rows # then shows every value in the table using color ranging from red (lowest) to white (highest) # Computation may take some time heatmap(as.matrix(a),Colv=NA) # Previous heatmap normalized each row, the next one uses data as they are: heatmap(as.matrix(a),Colv=NA,scale="none")
# k means clustering to 7 clusters k=7 cl <- kmeans(a,k) # each gene has assigned a cluster (number between 1 and k) cl$cluster # draw only cluster number 3 out of k heatmap(as.matrix(a[cl$cluster==3,]),Rowv=NA, Colv=NA) # reorder genes in the table according to cluster heatmap(as.matrix(a[order(cl$cluster),]),Rowv=NA, Colv=NA) # compare overall column means with column means in cluster 3 # function apply uses mean on every column (or row if 2 changed to 1) apply(a,2,mean) # now means within cluster apply(a[cl$cluster==3,],2,mean) # clusters have centers which are also computed as means # so this is the same as previous command cl$centers[3,]
Task D
- Draw a plot in which x-axis is time and y-axis is the expression level and the center of each cluster is shown as a line
- use command matplot(x,y,type="l") which gets two matrices x and y and plots columns of one vs the other
- matplot(,y,type="l") will use numbers 1,2,3... as columns of the missing matrix x
- create y from cl$centers by applying function t (transpose)
- to create an appropriate matrix x, create a vector of times for individual experiments in minutes or hours (do it manually, no need to parse column names automatically)
- using functions rep and matrix you can create a matrix x in which this vector is used as every column
- then run matplot(x,y,type="l")
- since time points are not evenly spaced, it would be better to use logscale: matplot(x,y,type="l",log="x")
- to avoid log(0), change the first timepoint from 0min to 1min
- Submit file clusters.png with your final plot
L06
Topic of this lecture are statistical tests in R.
- Beginners in statistics: listen to lecture, then do tasks A, B, C
- If you know basics of statistical tests, do tasks B, C, D
- More information on this topic in 1-EFM-340 Počítačová štatistika
Introduction to statistical tests: sign test
- [18]
- Two friends A and B have played their favourite game n=10 times, A has won 6 times and B has won 4 times.
- A claims that he is a better player, B claims that such a result could easily happen by chance if they were equally good players.
- Hypothesis of player B is called null hypothesis that the pattern we see (A won more often than B) is simply a result of chance
- Null hypothesis reformulated: we toss coin n times and compute value X: the number of times we see head. The tosses are independent and each toss has equal probability of being 0 or 1
- Similar situation: comparing programs A and B on several inputs, counting how many times is program A better than B.
# simulation in R: generate 10 psedorandom bits
# (1=player A won)
sample(c(0,1), 10, replace = TRUE)
# result e.g. 0 0 0 0 1 0 1 1 0 0
# directly compute random variable X, i.e. sum of bits
sum(sample(c(0,1), 10, replace = TRUE))
# result e.g. 5
# we define a function which will m times repeat
# the coin tossing experiment with n tosses
# and returns a vector with m values of random variable X
experiment <- function(m, n) {
x = rep(0, m) # create vector with m zeroes
for(i in 1:m) { # for loop through m experiments
x[i] = sum(sample(c(0,1), n, replace = TRUE))
}
return(x) # return array of values
}
# call the function for m=20 experiments, each with n tosses
experiment(20,10)
# result e.g. 4 5 3 6 2 3 5 5 3 4 5 5 6 6 6 5 6 6 6 4
# draw histograms for 20 experiments and 1000 experiments
png("hist10.png") # open png file
par(mfrow=c(2,1)) # matrix of plots with 2 rows and 1 column
hist(experiment(20,10))
hist(experiment(1000,10))
dev.off() # finish writing to file
- It is easy to realize that we get binomial distribution (binomické rozdelenie)
- P-value of the test is the probability that simply by chance we would get k the same or more extreme than in our data.
- In other words, what is the probability that in 10 tosses we see head 6 times or more (one sided test)
- If the p-value is very small, say smaller than 0.01, we reject the null hypothesis and assume that player A is in fact better than B
# computing the probability that we get exactly 6 heads in 10 tosses
dbinom(6, 10, 0.5) # result 0.2050781
# we get the same as our formula above:
7*8*9*10/(2*3*4*(2^10)) # result 0.2050781
# entire probability distribution: probabilities 0..10 heads in 10 tosses
dbinom(0:10, 10, 0.5)
# [1] 0.0009765625 0.0097656250 0.0439453125 0.1171875000 0.2050781250
# [6] 0.2460937500 0.2050781250 0.1171875000 0.0439453125 0.0097656250
# [11] 0.0009765625
#we can also plot the distribution
plot(0:10, dbinom(0:10, 10, 0.5))
barplot(dbinom(0:10,10,0.5))
#our p-value is sum for 7,8,9,10
sum(dbinom(6:10,10,0.5))
# result: 0.3769531
# so results this "extreme" are not rare by chance,
# they happen in about 38% of cases
# R can compute the sum for us using pbinom
# this considers all values greater than 5
pbinom(5, 10, 0.5, lower.tail=FALSE)
# result again 0.3769531
# if probability is too small, use log of it
pbinom(9999, 10000, 0.5, lower.tail=FALSE, log.p = TRUE)
# [1] -6931.472
# the probability of getting 10000x head is exp(-6931.472) = 2^{-100000}
# generating numbers from binomial distribution
# - similarly to our function experiment
rbinom(20, 10, 0.5)
# [1] 4 4 8 2 6 6 3 5 5 5 5 6 6 2 7 6 4 6 6 5
# running the test
binom.test(6, 10, p = 0.5, alternative="greater")
#
# Exact binomial test
#
# data: 6 and 10
# number of successes = 6, number of trials = 10, p-value = 0.377
# alternative hypothesis: true probability of success is greater than 0.5
# 95 percent confidence interval:
# 0.3035372 1.0000000
# sample estimates:
# probability of success
# 0.6
# to only get p-value run
binom.test(6, 10, p = 0.5, alternative="greater")$p.value
# result 0.3769531
Comparing two sets of values: Welch's t-test
- Let us now consider two sets of values drawn from two normal distributions with unknown means and variances
- The null hypothesis of the Welch's t-test is that the two distributions have equal means
- The test computes test statistics (in R for vectors x1, x2):
- (mean(x1)-mean(x2))/sqrt(var(x1)/length(x1)+var(x2)/length(x2))
- This test statistics is approximately distributed according to Student's t-distribution with the degree of freedom obtained by
n1=length(x1) n2=length(x2) (var(x1)/n1+var(x2)/n2)**2/(var(x1)**2/((n1-1)*n1*n1)+var(x2)**2/((n2-1)*n2*n2))
- Luckily R will compute the test for us simply by calling t.test
x1 = rnorm(6, 2, 1) # 2.70110750 3.45304366 -0.02696629 2.86020145 2.37496993 2.27073550 x2 = rnorm(4, 3, 0.5) # 3.258643 3.731206 2.868478 2.239788 > t.test(x1,x2) # t = -1.2898, df = 7.774, p-value = 0.2341 # alternative hypothesis: true difference in means is not equal to 0 # means 2.272182 3.024529 x2 = rnorm(4, 5, 0.5) # 4.882395 4.423485 4.646700 4.515626 t.test(x1,x2) # t = -4.684, df = 5.405, p-value = 0.004435 # means 2.272182 4.617051 # to get only p-value, run t.test(x1,x2)$p.value
We will apply Welch's t-test to microarray data
- Data from GEO database [19], publication [20]
- Abbott et al 2007: Generic and specific transcriptional responses to different weak organic acids in anaerobic chemostat cultures of Saccharomyces cerevisiae
- gene expression measurements under 5 conditions:
- reference: yeast grown in normal environment
- 4 different acids added so that cells grow 50% slower (acetic, propionic, sorbic, benzoic)
- from each condition (reference and each acid) we have 3 replicates
- together our table has 15 columns (3 replicates from 5 conditions)
- 6398 rows (genes)
- We will test statistical difference between the reference condition and one of the acids (3 numbers vs other 2 numbers)
- See Task B in #HW06
Multiple testing correction
- When we run t-tests on the reference vs. acetic acid on all 6398 genes, we get 118 genes with p-value<=0.01
- Purely by chance this would happen in 1% of cases (from definition of p-value)
- So purely by chance we would expect to get about 64 genes with p-value<=0.01
- So perhaps roughly half of our detected genes (maybe less, maybe more) are false positives
- Sometimes false positives may even overwhelm results
- Multiple testing correction tries to limit the number of false positives among results of multiple statistical tests
- Many different methods
- The simplest one is Bonferroni correction, where the threshold on p-value is divided by the number of tested genes, so instead of 0.01 we use 0.01/6398 = 1.56e-6
- This way the expected overall number of false positives in the whole set is 0.01 and so the probability of getting even a single false positive is also at most 0.01 (by Markov inequality)
- We could instead multiply all p-values by the number of tests and apply the original threshold 0.01 - such artificially modified p-values are called corrected
- After Bonferroni correction we get only 1 significant gene
# the results of p-tests are in vector pa of length 6398 # manually multiply p-values by length(pa), count those that have value <=0.01 sum(pa * length(pa) < 0.01) # in R you can use p.adjust form multiple testing correction pa.adjusted = p.adjust(pa, method ="bonferroni") # this is equivalent to multiplying by the length and using 1 if the result > 1 pa.adjusted = pmin(pa*length(pa),rep(1,length(pa))) # there are less conservative multiple testing correction methods, e.g. Holm's method # but in this case we get almost the same results pa.adjusted2 = p.adjust(pa, method ="holm")
- Other frequently used correction is false discovery rate (FDR), which is less strict and controls the overall proportion of false positives among results
HW06
- Do either tasks A,B,C (beginners) or B,C,D (more advanced). If you really want, you can do all four for bonus credit.
- In your protocol write used R commands with brief comments on your approach.
- Submit required plots with filenames as specified.
- For each task also include results as required and a short discussion commenting the results/plots you have obtained. Is the value of interest increasing or decreasing with some parameter? Are the results as expected or surprising?
- Outline of protocol is in /tasks/hw06/protocol.txt
Task A: sign test
- Consider a situation in which players played n games, out of which a fraction of q were won by A (example in lecture corresponds to q=0.6 and n=10)
- Compute a table of p-values for n=10,20,...,90,100 and for q=0.6, 0.7, 0.8, 0.9
- Plot the table using matplot (n is x-axis, one line for each value of q)
- Submit the plot in sign.png
- Discuss the values you have seen in the plot / table
Outline of the code:
# create vector rows with values 10,20,...,100
rows=(1:10)*10
# create vector columns with required values of q
columns=c(0.6, 0.7, 0.8, 0.9)
# create empty matrix of pvalues
pvalues = matrix(0,length(rows),length(columns))
# TODO: fill in matrix pvalues using binom.test
# set names of rows and columns
rownames(pvalues)=rows
colnames(pvalues)=columns
# careful: pvalues[10,] is now 10th row, i.e. value for n=100,
# pvalues["10",] is the first row, i.e. value for n=10
# check that for n=10 and q=0.6 you get p-value 0.3769531
pvalues["10","0.6"]
# create x-axis matrix (as in HW05, part D)
x=matrix(rep(rows,length(columns)),nrow=length(rows))
# matplot command
png("sign.png")
matplot(x,pvalues,type="l",col=c(1:length(columns)),lty=1)
legend("topright",legend=columns,col=c(1:length(columns)),lty=1)
dev.off()
Task B: Welch's t-test on microarray data
- Read table with microarray data, transform it to log scale, then work with table a:
input=read.table("/tasks/hw06/acids.tsv", header=TRUE, row.names=1)
a = log(input)
- Columns 1,2,3 are reference, columns 4,5,6 acetic acid, 7,8,9 benzoate, 10,11,12 propionate, and 13,14,15 sorbate
- Write a function my.test which will take as arguments table a and 2 lists of columns (e.g. 1:3 and 4:6) and will run for each row of the table Welch's t-test of the first set of columns vs the second set. It will return the resulting vector of p-values
- For example by calling pa <- my.test(a, 1:3, 4:6) we will compute p-values for differences between reference and acetic acids (computation may take some time)
- The first 5 values of pa should be
> pa[1:5] [1] 0.94898907 0.07179619 0.24797684 0.48204100 0.23177496
- Run the test for all four acids
- Report how many genes were significant with p-value <= 0.01 for each acid
- See Vector arithmetics in HW05
- You can count TRUE items in a vector of booleans by sum, e.g. sum(TRUE,FALSE,TRUE) is 2
- Report how many genes are significant for both acetic and benzoate acids? (logical and is written as &)
Task C: multiple testing correction
Run the following snippet of code, which works on the vector of p-values pa obtained for acetate in task B
# adjusts vectors of p-vales from tasks B for using Bonferroni correction pa.adjusted = p.adjust(pa, method ="bonferroni") # add this adjusted vector to frame a a <- cbind(a, pa.adjusted) # create permutation ordered by pa.adjusted oa = order(pa.adjusted) # select from table five rows with the lowest pa.adjusted (using vector oa) # and display columns containing reference, acetate and adjusted p-value a[oa[1:5],c(1:6,16)]
You should get output like this:
ref1 ref2 ref3 acetate1 acetate2 acetate3 pa.adjusted SUL1 7.581312 7.394985 7.412040 2.1633230 2.05412373 1.9169226 0.004793318 YMR244W 2.985682 2.975530 3.054001 0.3364722 0.33647224 0.1823216 0.188582576 DIP5 6.943991 7.147795 7.296955 0.6931472 0.09531018 0.5306283 0.253995075 YLR460C 5.620401 5.801212 5.502482 3.2425924 3.48431229 3.3843903 0.307639012 HXT4 2.821379 3.049273 2.772589 7.7893717 8.24446541 8.3041980 0.573813502
Do the same procedure for benzoate p-values and report the result. Comment the results for both acids.
Task D: volcano plot, test on data generated from null hypothesis
Draw a volcano plot for the acetate data
- x-axis of this plot is the difference in the mean of reference and mean of acetate.
- You can compute row means of a matrix by rowMeans.
- y-axis is -log10 of the p-value (use original p-values before multiple testing correction)
- You can quickly see genes which have low p-values (high on y-axis) and also big difference in mean expression between the two conditions (far from 0 on x-axis). You can also see if acetate increases or decreases expression of these genes.
Now create a simulated dataset sharing some features of the real data but observing the null hypothesis that the mean of reference and acetate are the same for each gene
- Compute vector m of means for columns 1:6 from matrix a
- Compute vectors sr and sa of standard deviations for reference columns and for acetate columns respectively
- You can compute standard deviation for each row of a matrix by apply(some.matrix, 1, sd)
- For each for each i in 1:6398, create three samples from normal distribution with mean m[i] and standard deviation sr[i], and three samples with mean m[i] and deviation sa[i]
- Use function rnorm
- On the resulting matrix apply Welches' t-test and draw the volcano plot.
- How many random genes have p-value <=0.01? Is it roughly what we would expect under the null hypothesis?
Draw histogram of p-values from the real data (reference vs acetate) and from random data. Describe how they differ. Is it what you would expect?
- use function hist
Submit plots volcano-real.png, volcano-random.png, hist-real.png, hist-random.png (real for real expression data and random for generated data)
L07
In this lecture we dive into SQLite3 and Python.
SQLite3
SQLite3 is a simple "database" stored in one file. Think of SQLite not as a replacement for Oracle but as a replacement for fopen(). Documentation: https://www.sqlite.org/docs.html
You can access sqlite database either from command line:
usamec@Darth-Labacus-2:~$ sqlite3 db.sqlite3 SQLite version 3.8.2 2013-12-06 14:53:30 Enter ".help" for instructions Enter SQL statements terminated with a ";" sqlite> CREATE TABLE test(id integer primary key, name text); sqlite> .schema test CREATE TABLE test(id integer primary key, name text); sqlite> .exit
Or from python interface: https://docs.python.org/2/library/sqlite3.html.
Python
Python is a perfect language for almost anything. Here is a cheatsheet: http://www.cogsci.rpi.edu/~destem/igd/python_cheat_sheet.pdf
Scraping webpages
The simplest tool for scraping webpages is urllib2: https://docs.python.org/2/library/urllib2.html Example usage:
import urllib2
f = urllib2.urlopen('http://www.python.org/')
print f.read()
Parsing webpages
We use beautifulsoup4 for parsing html (http://www.crummy.com/software/BeautifulSoup/bs4/doc/). I recommend following examples at the beginning of the documentation and example about CSS selectors: http://www.crummy.com/software/BeautifulSoup/bs4/doc/#css-selectors
Parsing dates
You have two options. Either use datetime.strptime or use dateutil package (https://dateutil.readthedocs.org/en/latest/parser.html).
Other usefull tips
- Don't forget to commit to your sqlite3 database (db.commit()).
- CREATE TABLE IF NOT EXISTS can be usefull at the start of your script.
- Inspect element (right click on element) in Chrome can be very helpful.
- Use screen command for long running scripts.
- All packages are installed on vyuka server. If you are planning using your own laptop, you need to install them using pip (preferably using virtualenv).
HW07
- Submit by copying requested files to /submit/hw07/username/
General goal: Scrape comments from several (hundreds) sme.sk users from last month and store them in SQLite3 database.
Task A
Create SQLite3 "database" with appropriate schema for storing comments from SME.sk discussions. You will probably need tables for users and comments. You don't need to store which comments replies to which one.
Submit two files:
- db.sqlite3 - the database
- schema.txt - brief description of your schema and rationale behind it
Task B
Build a crawler, which crawls comments in sme.sk discussions. You have two options:
- For fewer points: Script which gets url of the user (http://ekonomika.sme.sk/diskusie/user_profile.php?id_user=157432) and crawls his comments from last month.
- For more points: Scripts which gets one starting url (either user profile or some discussion, your choice) and automatically discovers users and crawls their comments.
This crawler should store comments in SQLite3 database built in previous task. Submit following:
- db.sqlite3 - the database
- every python script used for crawling
- README (how to start your crawler)
L08
In this lecture we will use Flask and simple text processing utilities from ScikitLearn.
Flask
Flask is simple web server for python (http://flask.pocoo.org/docs/0.10/quickstart/#a-minimal-application) You can find sample flask application at /tasks/hw08/simple_flask. Before running change the port number. You can then access your app at vyuka.compbio.fmph.uniba.sk:4247 (change port number).
There may be problem with access to strange port numbers due to firewalling rules. There are at least two ways to circumvent this:
- Use X forwarding and run web browser directly from vyuka
local_machine> ssh vyuka.compbio.fmph.uniba.sk -XC vyuka> chromium-browser
- Create SOCKS proxy to vyuka.compbio.fmph.uniba.sk and set SOCKS proxy at that port on your local machine. Then all web traffic goes through vyuka.compbio.fmph.uniba.sk via ssh tunnel. To create SOCKS proxy server on local machine port 8000 to vyuka.compbio.fmph.uniba.sk:
local_machine> ssh vyuka.compbio.fmph.uniba.sk -D 8000
(keep ssh session open while working)
Flask uses jinja2 (http://jinja.pocoo.org/docs/dev/templates/) templating language for showing html (you can use strings in python but it is painful).
Processing text
Main tool for processing text is CountVectorizer class from ScikitLearn (http://scikit--learn.org/stable/modules/generated/sklearn.feature_extraction.text.CountVectorizer.html). It transforms text into bag of words (for each word we get counts). Example:
from sklearn.feature_extraction.text import CountVectorizer vec = CountVectorizer(strip_accents='unicode') texts = [ "Ema ma mamu.", "Zirafa sa vo vani kupe a hneva sa." ] t = vec.fit_transform(texts).todense() print t print vec.vocabulary_
Useful things
We are working with numpy arrays here (that's array t in example above) Numpy arrays has also lots of nice tricks. First lets create two matrices:
>>> import numpy as np
>>> a = np.array([[#1,2,3],[4,5,6]])
>>> b = np.array([[#7,8],[9,10],[11,12]])
>>> a
array([[#1, 2, 3],
[4, 5, 6]])
>>> b
array([[# 7, 8],
[ 9, 10],
[11, 12]])
We can sum this matrices or multiply them by some number:
>>> 3 * a
array([[# 3, 6, 9],
[12, 15, 18]])
>>> a + 3 * a
array([[# 4, 8, 12],
[16, 20, 24]])
We can calculate sum of elements in each matrix, or sum by some axis:
>>> np.sum(a) 21 >>> np.sum(a, axis=1) array([ 6, 15]) >>> np.sum(a, axis=0) array([5, 7, 9])
There is a lot other usefull functions check https://docs.scipy.org/doc/numpy-dev/user/quickstart.html.
This can help you get top words for each user: http://docs.scipy.org/doc/numpy/reference/generated/numpy.argsort.html#numpy.argsort
HW08
- Submit by copying requested files to /submit/hw08/username/
General goal: Build a simple website, which lists all crawled users and for each users has a page with simple statistics for given user.
This lesson requires crawled data from previous lesson, if you don't have one, you can find it at (and thank Baska): /tasks/hw08/db.sqlite3
Submit source code (web server and preprocessing scripts) and database files.
Task A
Create a simple flask web application which:
- Has a homepage where is a list of all users (with links to their pages).
- Has a page for each user, which has simple information about user: His nickname, number of posts and hist last 10 posts.
Task B
For each user preprocess and store list of his top 10 words and list of top 10 words typical for him (which he uses much more often than other users, come up with some simple heuristics). Show this information on his page.
Task C
Preprocess and store list of top three similar users for each user (try to come up with some simple definition of similarity based on text in posts). Again show this information on user page.
Bonus: Try to use some simple topic modeling (e.g. PCA as in TruncatedSVD from scikit-learn) and use it for finding similar users.
L09
In this lesson we make simple javascript visualizations.
Your goal is to take examples from here https://developers.google.com/chart/interactive/docs/ and tweak them for your purposes.
Tips:
- You can output your data into javascript data structures in Flask template. It is a bad practice, but sufficient for this lesson. (Better way is to load JSON through API).
- Remember that you have to bypass the firewall.
HW09
- Submit by copying requested files to /submit/hw09/username/
General goal: Extend user pages from previous project with simple visualizations.
Task A
Show a calendar, which shows during which days was user active (like this https://developers.google.com/chart/interactive/docs/gallery/calendar#overview).
Task B
Show a histogram of comments length (like this https://developers.google.com/chart/interactive/docs/gallery/histogram#example).
Task C
Try showing a word tree for a user (https://developers.google.com/chart/interactive/docs/gallery/wordtree#overview). Try to normalize the text (lowercase, remove accents). CountVectorizer has method build_analyzer, which returns a function, which does this for you.
L10
Hadoop is a distributed filesystem and a map/reduce framework. It allows large data sets to be stored across multiple computers and also efficiently processed through the map reduce paradigm.
Hadoop setup
In order to use haddop on the server, you must source a script that sets up all the required environment variables needed to run hadoop:
# set up hadoop environment source ~yoyo/hadoop/hadoop-env.sh
After that you should be able to start the hadoop command.
Distributed filesystem
In order to process any data it must be stored in hadoop's distributed filesystem (HDFS). Hadoop exposes the usual filesystem manipulation commands:
# hadoop filesystem with usual commands hadoop fs -ls hadoop fs -ls / hadoop fs -mkdir input hadoop fs -cp /input/example/pg100.txt input/ hadoop fs -ls input
Note that paths beginning with / are absolute, without slash are relative to
your home directory which is /user/USERNAME.
To transfer files between the server and hadoop filesystems you can use the
put and get (you can also use the cat
command to get contents of short text files).
hadoop fs -put someLocalFile.txt input/ hadoop fs -get output/part-00000 .
Map Reduce
Map/reduce is a programming model / framework that allows processing large ammounts of (distributed) data in a distributed manner.
The system resembles the map nad reduce functions.
Input data (files) is first processed by the map function into key-value pairs,
these are redistributed so that all pairs with the same key are together and
and then processed by the reduce function to produce the final result.
Multiple "mappers" and "reducers" can be run on different machines (i.e. mappers can be run directly on the nodes where the data resides and thus no copying of data needs to be done).
Simple word count example
- Given a lot of large files distributed over several servers, produce word statistics: for each word how many times it occurs in the files.
- map: for each input file count the occurrence of each word in this file
- the result is a list of items separated by tab: word number
- hadoop will group together map results from all partial files and will put the same keys together
- reduce will sum together adjacent values with each key
Map part
- read file word by word
- for each word from input print this word, tab and 1 (hadoop will group the same words together anyway)
echo "abc abc def abc " | ~yoyo/example/map.awk
Redistribute / shuffle
Hadoop wil collect together the same keys, i.e. the result is something like this:
echo "abc abc def abc " | ~yoyo/example/map.awk | sort
Reduce part
Reducer then does something similar to niq -c, i.e. counts adajcent equal words. Here implemented in bash
echo "abc abc def abc " | ~yoyo/example/map.awk | sort | ~yoyo/example/reduce.sh
Running in hadoop
Normallly Hadoop Map-Reduce transformations are written in Java. However the default installation of hadoop contains a map-reduce class (hadoop streaming) that cat execute any process and write input to / read output from it.
Another technical problem with this solution is that the executed programs need to be present on all nodes where
the mappers / reducers will be run. They either can be installed on every node or present in the haddop filesystem.
The streaming class has an option (-file) to automatically upload any progra / data files along with the job.
# the streaming library
JAR=${HADOOP_PREFIX}/share/hadoop/tools/lib/hadoop-streaming-2.7.3.jar
BINKS=""
# use files in the /input/example directory for input
# place output in the out directory
# use map.awk and reduce.sh as mapper / reducer, upload them to haddop fs along the job
hadoop jar $JAR $BINKS -mapper map.awk -reducer reduce.sh -input /input/example/ -output out -file map.awk -file reduce.sh
HW10
- Submit the protocol, scripts and required output files.
- Before running the
hadoopcommand, source a script the necessary settings / paths:
source ~yoyo/hadoop/hadoop-env.sh
- The directory
/input/portage/logscontains large ammounts of log files from a gentoo instalation. - Installation of files is recorded in the logs as follows:
--- /usr/bin/ >>> /usr/bin/make -> gmake >>> /usr/bin/gmake
The first line represents a directory, the second records a symlink being created (we want to include these in the list of files), the third line represents a regular file being installed.
Task A
- Find out names of all the files that were ever installed and put them to a file
installed_all.txt.
Task B
- Find out 10 most rewriten / reinstalled files (you can use
sort/headto postprocess the results file). Write the results (most reinstalled file first) to the filetop_reinstalled.txt.
Task C
- For each package find the number of errors that ever happened during its installation. Write the results to file
package_errors.txtin the formpackage_name numErrors, sorted with package with most errors on the top. - Treat any occurrence of the string
error:as an error (including the colon, case insensitive) - Packages are marked at the start of each log file with a line (note the space at the start of the line):
* Package: sys-devel/make-4.1-r1
L11
Biological story: tiny monkeys
- Common marmoset (Callithrix jacchus, Kosmáč bielofúzý) weights only about 1/4 kg
- Most primates are much bigger
- Which marmoset genes differ from other primates and are related to the small size?
- Positive selection scan computes of each gene a p-value, whether it evolved on the marmoset lineage faster
- The result is a list of p-values, one for each gene
- Which biological functions are enriched among positively selected genes? Are any of those functions possibly related to body size?
Gene functions and GO categories
Use mysql database "marmoset" on the server.
- We can look at the description of a particular gene:
select * from genes where prot='IGF1R'; +----------------------------+-------+-------------------------------------------------+ | transcriptid | prot | description | +----------------------------+-------+-------------------------------------------------+ | knownGene.uc010urq.1.1.inc | IGF1R | insulin-like growth factor 1 receptor precursor | +----------------------------+-------+-------------------------------------------------+
- In the database, we have stored all the P-values from positive selection tests:
select * from lrtmarmoset where transcriptid='knownGene.uc010urq.1.1.inc'; +----------------------------+---------------------+ | transcriptid | pval | +----------------------------+---------------------+ | knownGene.uc010urq.1.1.inc | 0.00142731425252827 | +----------------------------+---------------------+
- Genes are also assigned functional categories based on automated processes (including sequence similarity to other genes) and manual curation. The corresponding database is maintained by Gene Ontology Consortium. We can use on-line sources to search for these annotations, e.g. here.
- We can also download the whole database and preprocess it into usable form:
select * from genes2gocat,gocatdefs where transcriptid='knownGene.uc010urq.1.1.inc' and genes2gocat.cat=gocatdefs.cat; (results in 50 categories)
- GO categories have a hierarchical structure - see for example category GO:0005524 ATP binding:
select * from gocatparents,gocatdefs where gocatparents.parent=gocatdefs.cat and gocatparents.cat='GO:0005524'; +------------+------------+---------+------------+-------------------------------+ | cat | parent | reltype | cat | def | +------------+------------+---------+------------+-------------------------------+ | GO:0005524 | GO:0032559 | isa | GO:0032559 | adenyl ribonucleotide binding | +------------+------------+---------+------------+-------------------------------+ ... and continuing further up the hierarchy: | GO:0032559 | GO:0030554 | isa | GO:0030554 | adenyl nucleotide binding | | GO:0032559 | GO:0032555 | isa | GO:0032555 | purine ribonucleotide binding | | GO:0030554 | GO:0001883 | isa | GO:0001883 | purine nucleoside binding | | GO:0030554 | GO:0017076 | isa | GO:0017076 | purine nucleotide binding | | GO:0032555 | GO:0017076 | isa | GO:0017076 | purine nucleotide binding | | GO:0032555 | GO:0032553 | isa | GO:0032553 | ribonucleotide binding | | GO:0001883 | GO:0001882 | isa | GO:0001882 | nucleoside binding | | GO:0017076 | GO:0000166 | isa | GO:0000166 | nucleotide binding | | GO:0032553 | GO:0000166 | isa | GO:0000166 | nucleotide binding | | GO:0001882 | GO:0005488 | isa | GO:0005488 | binding | | GO:0000166 | GO:0005488 | isa | GO:0005488 | binding | | GO:0005488 | GO:0003674 | isa | GO:0003674 | molecular_function |
- What else can be under GO:0032559 adenyl ribonucleotide binding?
select * from gocatparents,gocatdefs where gocatparents.cat=gocatdefs.cat and gocatparents.parent='GO:0032559'; +------------+------------+---------+------------+-------------+ | cat | parent | reltype | cat | def | +------------+------------+---------+------------+-------------+ | GO:0005524 | GO:0032559 | isa | GO:0005524 | ATP binding | | GO:0016208 | GO:0032559 | isa | GO:0016208 | AMP binding | | GO:0043531 | GO:0032559 | isa | GO:0043531 | ADP binding | +------------+------------+---------+------------+-------------+
Mann–Whitney U test
- also known as Wilcoxon rank-sum test
- In Lecture 6, we have used Welch's t-test to test if one set of expression measurements for a gene are significantly different from the second set
- This test assumes that both sets come from normal (Gaussian) distributions with unknown parameters
- Mann-Whitney U test is called non-parametric, because it does not make this assumption
- The null hypothesis is that two sets of measurements were generated by the same unknown probability distribution
- Alternative hypothesis: for X from the first distribution and Y from the second P(X>Y) is not equal P(Y>X)
- We will use a one-side version of the alternative hypothesis: P(X>Y) > P(Y>X)
- Compute test statistics U:
- compare all pairs X, Y (X from first set, Y from second set)
- if X>Y, add 1 to U
- if X==Y, add 0.5
- For large sets, U is approximately normally distributed under the null hypothesis
How to use in R:
# generate 20 samples from exponential distrib. with mean 1 x = rexp(20, 1) # generate 30 samples from exponential distrib. with mean 1/2 y = rexp(30, 2) # test if values of x greater than y wilcox.test(x,y,alternative="greater") # W = 441, p-value = 0.002336 # alternative hypothesis: true location shift is greater than 0 # W is the U statistics above # now generate y twice from the same distrib. as x y = rexp(30, 1) wilcox.test(x,y,alternative="greater") # W = 364, p-value = 0.1053 # relatively small p-value (by chance) y = rexp(30, 1) wilcox.test(x,y,alternative="greater") # W = 301, p-value = 0.4961 # now much greater p-value
Another form of the test, potentially useful for HW:
- have a vector of values x, binary vector b indicating two classes: 0 and 1
- test if values marked by 0 are greater than values marked by 1
# generate 10 with mean 1, 30 with mean 1/2, 10 with mean 1 x = c(rexp(10,1),rexp(30,2),rexp(10,1)) # classes 10x0, 20x1, 10x0 b = c(rep(0,10),rep(1,30),rep(0,10)) wilcox.test(x~b,alternative="greater") # the same test by distributing into subvectors x0 and x1 for classes 0 and 1 x0 = x[b==0] x1 = x[b==1] wilcox.test(x0,x1,alternative="greater") # should be the same as above
HW11
- In this task, you can use a combination of any scripting languages (e.g. Perl, Python, R) but also SQL, command-line tools etc.
- Input is in a database
- Submit required text files (optionally also files with figures in bonus part E)
- Also submit any scripts you have written for this HW
- In the protocol, include shell commands you have run
- Outline of protocol is in /tasks/hw11/protocol.txt
Available data
- All data necessary for this task is available in the mysql database 'marmoset' on the server
- You will find password in /tasks/hw11/readme.txt
- You have read-only access to the 'marmoset' database
- For creating temporary tables, etc., you can use database 'temp_youruserid' (e.g. 'temp_mrkvicka54'), where you are allowed to create new tables and store data
- You can address tables in mysql even between databases: to start client with your writeable database as default location, use:
- mysql -p temp_mrkvicka54
- You can then access data in the table 'genes' in the database 'marmoset' simply by using 'marmoset.genes'
Getting data from database:
- If your want to get data from database to a tab-separated file, write a select query, run with -e, redirect output:
- mysql -p marmoset -e 'select transcriptid as id, pval from lrtmarmoset' > pvals.tsv
Task A: Find ancestors of each GO category
- Compute a table (in your temporary db or in a file) which contains all pairs category and its ancestor
- In table gocatparents you have pairs category, its parent, so you need a transitive closure over this relation
- SQL is not very good at this, you can try repeated joins until you find no more ancestors
- Alternatively, you can simply extract data from the database and process them in a language of your choice
- Submit file sample-anc.txt which contains the list of all ancestors of GO:0042773, one per line, in sorted order
- There should be 14 such ancestors, excluding this category itself; the first in sorted order is GO:0006091, the last is GO:0055114
Task B: Gene in/out data for each GO category
- Again consider category GO:0042773
- Create a list of all genes that occur in table lrtmarmoset
- for each such gene list three columns separated by tabs: its transcript id, p-value from lrtmarmoset, and an indicator 0/1
- the indicator is 1, if this gene occurs in GO:0042773 or one of its subcategories; 0 otherwise
- to find, which gene occur directly in GO:0042773, use table genes2gocat, subcategories can be found in your table from part A
- note that genes2gocat contains more genes, we will consder only genes from lrtmarmoset
- Submit this file sample-genes.tsv
- Overall, your table should have 13717 genes, out of which 28 have value 1 in the last column
- The first lines of this list (when sorted alphabetically) might look as follos:
ensGene.ENST00000043410.1.inc 1 0 ensGene.ENST00000158526.1.inc 0.483315913388483 0 ... ensGene.ENST00000456284.1 1 1 ...
- Note that in part C, you will need to run this process for each category in the database, so make it sufficiently automated
Task C: Run Man-Whitney U test for each GO category
- Run Man-Whitney U test for each non-trivial category
- Non-trivial categories are such that at least one of our genes (from lrtmarmoset) is in the category and at least one of our genes is not in the category
- You should test, if genes in a particular GO category have smaller p-values in positive selection than genes outside the category
- List of all categories can be obtained from gocatdefs, but not all of them are non-trivial (there are 12455 non-trivial categories)
- Submit file test.tsv in which each line contains two tab separated values:
- GO category id
- p-value from the test
- For partial points test at least the category GO:0042773 from parts A and B
Task D: Report significant categories
- Submit file report.tsv with 20 most significant GO categories (lowest p-values)
- For each category list its ID, p-value and description
- Order them from the most significant
- Descriptions are in table gocatdefs
- To your protocol, write any observations you can make
- Do any reported categories seem interesting to you?
- Are any reported categories likely related to each other based on their descriptions?
Task E (bonus): cluster significant categories
- Some categories in task D appear similar according to their name
- Try creating k-means or hierarchical clustering of categories
- Represent each category as a binary vector in which for each gene you have one bit indicating if it is in the category
- Thus categories with the same set of genes will have identical vectors
- Try to report results in an appropriate form (table, text, figure), discuss them in the protocol
Note
- In part C, we have done many statistical tests, resulting P-values should be corrected by multiple testing correction from Lecture 6
- This is not required in this homework, but should be done in a real study