Vybrané partie z dátových štruktúr
2-INF-237, LS 2016/17
Hešovanie: Rozdiel medzi revíziami
(→Bloom filters (1970)) |
(→Sources) |
||
| 33 medziľahlých revízií od jedného používateľa nie je zobrazených. | |||
| Riadok 1: | Riadok 1: | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ==Introduction to hashing== | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | * Universe U = {0,..., u-1} | |
| + | * Table of size m | ||
| + | * Hash function h : U -> {0,...,m-1} | ||
| + | * Let X be the set of elements currently in the hash table, n = |X| | ||
| + | * We will assume n = Theta(m) | ||
| − | === | + | ===Totally random hash function=== |
| + | * (a.k.a uniform hashing model) | ||
| + | * select hash value h(x) for each x in U uniformly independently | ||
| + | * not practical - storing this hash function requires u * lg m bits | ||
| + | * used for simplified analysis of hashing in ideal case | ||
| − | * a bit string of length | + | ===Universal family of hash functions=== |
| − | * insert(x): set bits h1 (x), . . . , hk (x) to 1. | + | * some set of functions H |
| − | * test if x in the set: compute h1 (y), . . . , hk (y) and check whether all these bits are 1 | + | * draw a fuction h uniformly randomly from H |
| + | * there is a constant c such that for any distinct x,y from U we have Pr(h(x) = h(y)) <= c/m | ||
| + | * probability goes over choices of h | ||
| + | |||
| + | Note | ||
| + | * for totally random hash function, this probability is exactly 1/m | ||
| + | |||
| + | Example of a universal family: | ||
| + | * choose a fixed prime p >= u | ||
| + | * H_p = { h_a | h_a(x) =(ax mod p) mod m), 1<=a<=p-1 } | ||
| + | * p-1 hash functions, parameter a chosen randomly | ||
| + | |||
| + | Proof of universality | ||
| + | * consider x!=y, both from U | ||
| + | * if they collide (ax mod p) mod m = (ay mod p) mod m | ||
| + | * let c = (ax mod p), d = (ay mod p) | ||
| + | * note that c,d in {0..p-1} | ||
| + | * also c!=d because otherwise a(x-y) is divisible by p and both a and |x-y| are numbers from {1,...,p-1} and p is prime | ||
| + | * since c mod m = d mod m, then c-d = qm where 0<|qm|<p | ||
| + | * there are <=2(p-1)/m choices of q where |qm|<p | ||
| + | * we get a(x-y) mod p = qm mod p | ||
| + | * this has exactly one solution a in {1...p-1} | ||
| + | ** if there were two solutions a1 and a2, then (a1-a2)(x-y) would be divisible by p, which is again not possible | ||
| + | * overall at most 2(p-1)/m choices of hash functions collide x and y | ||
| + | * out of p-1 all hash functions in H_p | ||
| + | * therefore probability of collision <=2/m | ||
| + | |||
| + | ===Hashing with chaining, using universal family of h.f.=== | ||
| + | * linked list (or vector) for each hash table slot | ||
| + | * let c_i be the length of chain i, let <math>C_x=c_{h(x)}</math> for x in U | ||
| + | * any element y from X s.t. x!=y maps to h(x) with probability <= c/m | ||
| + | * so E[C_x] <= 1 + n * c / m = O(1) | ||
| + | ** from linearity of expectation - sum over indicator variables for each y in X if it collides with x | ||
| + | ** +1 in case x is in X | ||
| + | * so expected time of any search, insert and delete is O(1) | ||
| + | ** again, expectation taken over random choice of h from universal H | ||
| + | * however, what is the expected length of the longest chain in the table? | ||
| + | ** O(1+ sqrt(n^2 / m)) = O(sqrt(n)) | ||
| + | ** see Brass, p. 382 | ||
| + | ** there is a matching lower bound | ||
| + | * for totally random hash function this is "only" O(log n / log log n) | ||
| + | * similar bound proved also for some more realistic families of hash functions | ||
| + | * so the worst-case query is on average close to logarithmic | ||
| + | |||
| + | References | ||
| + | * Textbook Brass 2008 Advanced data structures | ||
| + | * Lecture L10 Erik Demaine z MIT: http://courses.csail.mit.edu/6.851/spring12/lectures/ | ||
| + | |||
| + | ==Perfect hashing== | ||
| + | |||
| + | Static case | ||
| + | * Fredman, Komlos, Szemeredi 1984 | ||
| + | * avoid any collisions, O(1) search in a static set | ||
| + | * two-level scheme: use a universal hash function to hash to a table of size m = Theta(n) | ||
| + | * each bucket i with set of elements X_i of size c_i hashed to a second-level hash of size m_i = alpha c_i^2 for some constant alpha | ||
| + | * again use a universal hash function for each second-level hash | ||
| + | * expected total number of collisions in a second level for slot i: | ||
| + | ** sum_{x,y in X_i, x!=y} Pr(h(x) = h(y)) <= (c_i)^2 c/m_i = c/alpha = O(1) | ||
| + | ** here c is the constant from definition universal family | ||
| + | * with a sufficently large alpha this expected number of collisions is <=1/2 | ||
| + | ** e.g. for c = 2 set alpha = 4 | ||
| + | * then with probability >=1/2 no collisions by Markov inequality | ||
| + | ** number of collisions is a random variable Y number with possible values 0,1,2,... | ||
| + | ** if E[Y]<=1/2, Pr(Y>=1)<=1/2 and thus Pr(Y=0)>=1/2 | ||
| + | * when building a hash table if we get a collision, randomly sample another hash function | ||
| + | * in O(1) expected trials get a good hash function | ||
| + | * expected space: m + sum_i m_i^2 = Theta(m + sum_i c_i^2) | ||
| + | * sum_i (c_i^2) is the number of pairs (x,y) s.t. x,y in X and h(x)=h(y); include x=y | ||
| + | * <math>\sum_{x,y\in X} \Pr(h(x)=h(y)) = n + \sum_{x!=y} \Pr(h(x)=h(y)) = n + n^2 c / m = \Theta(n)</math> | ||
| + | * so expected space is linear | ||
| + | * can be made worst-case by repeating construction if memory too big | ||
| + | * expected construction time O(n) | ||
| + | |||
| + | Dynamic perfect hashing | ||
| + | * amortized vector-like tricks | ||
| + | * when a 2-nd level hash table gets too full, allocate a bigger one | ||
| + | * O(1) deterministic search | ||
| + | * O(1) amortized expectd update | ||
| + | * O(n) expected space | ||
| + | |||
| + | References | ||
| + | * Lecture L10 Erik Demaine z MIT: http://courses.csail.mit.edu/6.851/spring12/lectures/ | ||
| + | * Textbook Brass 2008 Advanced data structures | ||
| + | |||
| + | ==Bloom Filter (Bloom 1970)== | ||
| + | |||
| + | * supports insert ''x'', test if ''x'' is in the set | ||
| + | ** may give false positives, e.g. claim that ''x'' is in the set when it is not | ||
| + | ** false negatives do not occur | ||
| + | |||
| + | '''Algorithm''' | ||
| + | * a bit string B[0,...,m-1] of length m, and k hash functions hi : U -> {0, ..., m-1}. | ||
| + | * insert(x): set bits B[h1(x)], ..., B[hk(x)] to 1. | ||
| + | * test if x in the set: compute h1(y), ..., hk(y) and check whether all these bits are 1 | ||
** if yes, claim x is in the set, but possibility of error | ** if yes, claim x is in the set, but possibility of error | ||
** if no, answer no, surely true | ** if no, answer no, surely true | ||
| − | + | '''Bounding the error''' If hash functions are totally random and independent, the probability of error is approximately (1-exp(-nk/m))^k | |
| − | * | + | * in fact probability somewhat higher for smaller values |
| − | + | * proof of the bound later | |
| − | * lg( | + | * totally random hash functions impractical (need to store hash value for each element of the universe), but the assumption simplifies analysis |
| − | * compare to a hash table, which needs at | + | * if we set k = ln(2) m/n, get error <math>(1-\exp(-\ln(2)))^{\ln(2)m/n}</math>. |
| − | + | * <math>\exp(-\ln(2)) = 1/2</math> and thus we get error <math>2^{-\ln(2)m/n} = c^{-m/n}</math>, where <math>c = 2^{\ln(2)} \approx 1.62</math> | |
| − | Use of Bloom filters | + | * to get error rate p for some n, we need <math>m = n \log_c (1/p) = n \lg(1/p)/\lg(c) = n \lg(1/p)/\ln(2) = n \lg(1/p) \lg(e)</math> |
| + | * for 1% error, we need about m=10n bits of space and k=7 | ||
| + | * memory size and error rate are independent of the size of the universe U | ||
| + | * compare to a hash table, which needs at least to store data items themselves (e.g. in n lg u bits) | ||
| + | * if we used k=1 (Bloom filter with one hash function), we need m=n/ln(1/(1-p)) bits, which for p=0.01 is about 99.5n, about 10 times more than with 7 hash functions | ||
| + | |||
| + | '''Use of Bloom filters''' | ||
* e.g. an approximate index of a larger data structure on a disk - if x not in the filter, do not bother looking at the disk, but small number of false positives not a problem | * e.g. an approximate index of a larger data structure on a disk - if x not in the filter, do not bother looking at the disk, but small number of false positives not a problem | ||
| + | * Example: Google BigTable maps row label, column label and timestamp to a record, underlies many Google technologies. It uses Bloom filter to check if a combination of row and column label occur in a given fragment of the table | ||
| + | ** For details, see Chang, Fay, et al. "Bigtable: A distributed storage system for structured data." ACM Transactions on Computer Systems (TOCS) 26.2 (2008): 4. [http://static.usenix.org/events/osdi06/tech/chang/chang_html/?em_x=22] | ||
| + | * see also A. Z. Broder and M. Mitzenmacher. Network applications of Bloom filters: A survey. In Internet Math. Volume 1, Number 4 (2003), 485-509. [http://projecteuclid.org/download/pdf_1/euclid.im/1109191032] | ||
| − | Proof of | + | '''Proof of the bound''' |
| + | * probability that some B[i] is set to 1 by hj(x) is 1/m | ||
| + | * probability that B[i] is not set to 1 is therefore 1-1/m and since we use k independent hash functions, the probability that B[i] is not set to one by any of them is (1-1/m)^k | ||
| + | * if we insert n elements, each is hashed by each function independently of other elements (hash functions are random) and thus Pr(B[i]=0)=(1-1/m)^{nk} | ||
| + | * Pr(B[i]=1) = 1-Pr(B[i]=0) | ||
| + | * consider a new element y which is not in the set | ||
| + | * error occurs when B[hj(y)]==1 for all j=1..k | ||
| + | * assume for now that these events are independent for different j | ||
| + | * error then occurs with probability Pr(B[i]=1)^k = (1-(1-1/m)^{nk})^k | ||
| + | * recall that <math>\lim_{x\to\infty} (1-1/x)^x = 1/e</math> | ||
| + | * thus probability of error is approximately (1-exp(-nk/m))^k | ||
| − | References | + | However, Pr(B[i]=1) are not independent |
| + | * consider the following experiment: have 2 binary random variables Y1, Y2, each generated independently, uniformly (i.e. P(Y1=1) = 0.5) | ||
| + | * Let X1 = Ya and X2 = Yb where a and b are chosen independently uniformly from set {1,2} | ||
| + | * X1 and X2 are not independent, e.g. Pr(X2 = 1|X1 = 1) = 2/3, whereas Pr(X2=1) = 1/2. (check the constants!) | ||
| + | * note: X1 and X2 would be independent conditioned on the sum of Y1+Y2. | ||
| + | |||
| + | * In Mitzenmacher & Upfal (2005), pp. 109–111, 308. - less strict independence assumption | ||
| + | |||
| + | '''Exercise''' | ||
| + | Let us assume that we have separate Bloom filters for sets A and B with the same set k hash functions. How can we create Bloom filters for union and intersection? Will the result be different from filter created directly for the union or for the intersection? | ||
| + | |||
| + | '''Theory''' | ||
| + | * Bloom filter above use about 1.44 n lg(1/p) bits to achieve error rate p. There is lower bound of n lg(1/p) [Carter et al 1978], constant 1.44 can be improved to 1+o(1) using more complex data structures, which are then probably less practical | ||
| + | * Pagh, Anna, Rasmus Pagh, and S. Srinivasa Rao. "An optimal Bloom filter replacement." SODA 2005. [http://www.itu.dk/~annao/Publications/Filter.pdf] | ||
| + | * L. Carter, R. Floyd, J. Gill, G. Markowsky, and M. Wegman. Exact and approximate membership testers. STOC 1978, pages 59–65. [http://www.researchgate.net/publication/234830711_Exact_and_approximate_membership_testers/file/e0b4951d5595258a88.pdf] | ||
| + | * see also proof of lower bound in Broder and Mitzenmacher 2003 below | ||
| + | |||
| + | '''Counting Bloom filters''' | ||
| + | * support insert and delete | ||
| + | * each B[i] is a counter with b bits | ||
| + | * insert increases counters, delete decreases | ||
| + | * assume no overflows, or reserve largest value 2^b-1 as infinity, cannot be increased or decreased | ||
| + | * Fan, Li, et al. "Summary cache: A scalable wide-area web cache sharing protocol." ACM SIGCOMM Computer Communication Review. Vol. 28. No. 4. ACM, 1998. [https://www.cs.princeton.edu/courses/archive/spring05/cos598E/bib/p254-fan.pdf] | ||
| + | |||
| + | '''References''' | ||
* Bloom, Burton H. (1970), [http://dx.doi.org/10.1145%2F362686.362692 "Space/Time Trade-offs in Hash Coding with Allowable Errors"], Communications of the ACM 13 (7): 422–426 | * Bloom, Burton H. (1970), [http://dx.doi.org/10.1145%2F362686.362692 "Space/Time Trade-offs in Hash Coding with Allowable Errors"], Communications of the ACM 13 (7): 422–426 | ||
* Bloom filter. In Wikipedia, The Free Encyclopedia. Retrieved January 17, 2015, from http://en.wikipedia.org/w/index.php?title=Bloom_filter&oldid=641921171 | * Bloom filter. In Wikipedia, The Free Encyclopedia. Retrieved January 17, 2015, from http://en.wikipedia.org/w/index.php?title=Bloom_filter&oldid=641921171 | ||
| + | * A. Z. Broder and M. Mitzenmacher. Network applications of Bloom filters: A survey. In Internet Math. Volume 1, Number 4 (2003), 485-509. [http://projecteuclid.org/download/pdf_1/euclid.im/1109191032] | ||
| + | |||
| + | ==Locality Sensitive Hashing== | ||
| + | * todo: expand and according to slides | ||
| + | |||
| + | * typical hashing tables used for exact search: is x in set X? | ||
| + | * nearest neighbor(x): find y in set X minimizing d(x,y) for a given distance measure d | ||
| + | ** or given x and distance threshold r, find all y in X with d(x,y)<=r | ||
| + | * want: polynomial time preprocessing of X, polylogarithmic queries | ||
| + | * later in the course: geometric data structures for similar problems in R^2 or higher dimensions | ||
| + | * but time or space grows exponentially with dimension (alternative is linear search) - "curse of dimensionality" | ||
| + | * examples of high-dimensional objects: images, documents, etc. | ||
| + | * locality sensitive hashing (LHS) is a randomized data structure not guaranteed to find the best answer but better running time | ||
| + | |||
| + | Approximate near neighbour (r,c)-NN | ||
| + | * preprocess a set X of size n for given r,c | ||
| + | * query(x): | ||
| + | ** if there is y in X such that d(y,x)<=r, return z in X such that d(z,x)<=c*r | ||
| + | ** LHS returns such z with some probability at least f | ||
| + | |||
| + | Example for Hamming distance in binary strings of length d | ||
| + | * Hash each string using L hashing functions, each using O(log n) randomly chosen string positions (hidden constant depends on d, r, c), then rehash using some other hash function to tables of size O(n) or a single table of size O(nL) | ||
| + | * Given q, find it using all L hash function, check every collision, report if distance at most cr. If checked more than 3L collisions, stop. | ||
| + | * L = O(n^{1/c}) | ||
| + | * Space O(nL + nd), query time O(Ld+L(d/r)log n) | ||
| + | * Probability of failure less than some constant f < 1, can be boosted by repeating independently | ||
| + | |||
| + | |||
| + | ===Minimum hashing=== | ||
| + | * similarity of two sets measured as Jaccard index J(A,B) = |A intersect B | / | A union B | | ||
| + | * distance d(A,B) = 1-J(A,B) | ||
| + | * used e.g. for approximate duplicate document detection based on set of words occurring in the document | ||
| + | |||
| + | * choose a totally random hash function h | ||
| + | * minhash_h(A) = \min_{x\in A} h(x) | ||
| + | * Pr(minhash_h(A)=minhash_h(B)) = J(A,B) = 1-d(A,B) if using totally random hash function an no collisions in A union B | ||
| + | ** where can be minimum in union? if in intersection, we get equality, otherwise not | ||
| + | * (r_1, r_2, 1-r_1, 1-r_2)-sensitive family (if we assume no collisions) | ||
| + | * again can we apply amplification | ||
| + | |||
| + | ===Notes=== | ||
| + | * Real-valued vectors: map to bits using random projections sgn (w · x + b) | ||
| + | * '''Data-dependent hashing:''' Use PCA, optimization, kernel tricks, even neural networks to learn hash functions good for a particular set of points and distance measure (or learn even distance measure from examples). [https://arxiv.org/pdf/1509.05472.pdf] | ||
| + | |||
| + | ===Sources=== | ||
| − | + | * Har-Peled, Sariel, Piotr Indyk, and Rajeev Motwani. "Approximate Nearest Neighbor: Towards Removing the Curse of Dimensionality." Theory of Computing 8.1 (2012): 321-350. [http://tocmirror.cs.tau.ac.il/articles/v008a014/v008a014.pdf] | |
| − | * Har-Peled, Sariel, Piotr Indyk, and Rajeev Motwani. "Approximate Nearest Neighbor: Towards Removing the Curse of Dimensionality." Theory of Computing 8.1 (2012): 321-350. | + | ** Indyk and Motwani: Approximate nearest neighbors: Towards removing the curse of dimensionality. In Proc. 30th STOC, pp. 604–613. ACM Press, 1998. |
| − | ** | + | * http://www.mit.edu/~andoni/LSH/ Pointers to papers |
| − | * | + | * Wikipedia |
| − | ** | + | ** http://en.wikipedia.org/wiki/Locality-sensitive_hashing |
| − | ** | + | ** http://en.wikipedia.org/wiki/MinHash |
| − | ** | + | * Several lectures: |
| − | ** | + | ** http://infolab.stanford.edu/~ullman/mining/2009/similarity3.pdf various distances, connection to minhashing |
| − | * | + | ** https://users.soe.ucsc.edu/~niejiazhong/slides/kumar.pdf nic intro, more duplicate detection, NN at the end |
| − | ** | + | ** http://delab.csd.auth.gr/courses/c_mmdb/mmdb-2011-2012-lsh2.pdf also nice intro through Hamming |
| + | * Charikar, Moses S. "Similarity estimation techniques from rounding algorithms." STOC 2002 [http://www.cs.princeton.edu/courses/archive/spring04/cos598B/bib/CharikarEstim.pdf] | ||
| + | ** simHash (cosine measure) | ||
Aktuálna revízia z 08:42, 11. apríl 2017
Obsah
Introduction to hashing
- Universe U = {0,..., u-1}
- Table of size m
- Hash function h : U -> {0,...,m-1}
- Let X be the set of elements currently in the hash table, n = |X|
- We will assume n = Theta(m)
Totally random hash function
- (a.k.a uniform hashing model)
- select hash value h(x) for each x in U uniformly independently
- not practical - storing this hash function requires u * lg m bits
- used for simplified analysis of hashing in ideal case
Universal family of hash functions
- some set of functions H
- draw a fuction h uniformly randomly from H
- there is a constant c such that for any distinct x,y from U we have Pr(h(x) = h(y)) <= c/m
- probability goes over choices of h
Note
- for totally random hash function, this probability is exactly 1/m
Example of a universal family:
- choose a fixed prime p >= u
- H_p = { h_a | h_a(x) =(ax mod p) mod m), 1<=a<=p-1 }
- p-1 hash functions, parameter a chosen randomly
Proof of universality
- consider x!=y, both from U
- if they collide (ax mod p) mod m = (ay mod p) mod m
- let c = (ax mod p), d = (ay mod p)
- note that c,d in {0..p-1}
- also c!=d because otherwise a(x-y) is divisible by p and both a and |x-y| are numbers from {1,...,p-1} and p is prime
- since c mod m = d mod m, then c-d = qm where 0<|qm|<p
- there are <=2(p-1)/m choices of q where |qm|<p
- we get a(x-y) mod p = qm mod p
- this has exactly one solution a in {1...p-1}
- if there were two solutions a1 and a2, then (a1-a2)(x-y) would be divisible by p, which is again not possible
- overall at most 2(p-1)/m choices of hash functions collide x and y
- out of p-1 all hash functions in H_p
- therefore probability of collision <=2/m
Hashing with chaining, using universal family of h.f.
- linked list (or vector) for each hash table slot
- let c_i be the length of chain i, let
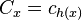 for x in U
for x in U
- any element y from X s.t. x!=y maps to h(x) with probability <= c/m
- so E[C_x] <= 1 + n * c / m = O(1)
- from linearity of expectation - sum over indicator variables for each y in X if it collides with x
- +1 in case x is in X
- so expected time of any search, insert and delete is O(1)
- again, expectation taken over random choice of h from universal H
- however, what is the expected length of the longest chain in the table?
- O(1+ sqrt(n^2 / m)) = O(sqrt(n))
- see Brass, p. 382
- there is a matching lower bound
- for totally random hash function this is "only" O(log n / log log n)
- similar bound proved also for some more realistic families of hash functions
- so the worst-case query is on average close to logarithmic
References
- Textbook Brass 2008 Advanced data structures
- Lecture L10 Erik Demaine z MIT: http://courses.csail.mit.edu/6.851/spring12/lectures/
Perfect hashing
Static case
- Fredman, Komlos, Szemeredi 1984
- avoid any collisions, O(1) search in a static set
- two-level scheme: use a universal hash function to hash to a table of size m = Theta(n)
- each bucket i with set of elements X_i of size c_i hashed to a second-level hash of size m_i = alpha c_i^2 for some constant alpha
- again use a universal hash function for each second-level hash
- expected total number of collisions in a second level for slot i:
- sum_{x,y in X_i, x!=y} Pr(h(x) = h(y)) <= (c_i)^2 c/m_i = c/alpha = O(1)
- here c is the constant from definition universal family
- with a sufficently large alpha this expected number of collisions is <=1/2
- e.g. for c = 2 set alpha = 4
- then with probability >=1/2 no collisions by Markov inequality
- number of collisions is a random variable Y number with possible values 0,1,2,...
- if E[Y]<=1/2, Pr(Y>=1)<=1/2 and thus Pr(Y=0)>=1/2
- when building a hash table if we get a collision, randomly sample another hash function
- in O(1) expected trials get a good hash function
- expected space: m + sum_i m_i^2 = Theta(m + sum_i c_i^2)
- sum_i (c_i^2) is the number of pairs (x,y) s.t. x,y in X and h(x)=h(y); include x=y
-
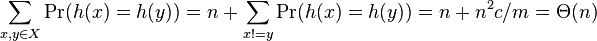
- so expected space is linear
- can be made worst-case by repeating construction if memory too big
- expected construction time O(n)
Dynamic perfect hashing
- amortized vector-like tricks
- when a 2-nd level hash table gets too full, allocate a bigger one
- O(1) deterministic search
- O(1) amortized expectd update
- O(n) expected space
References
- Lecture L10 Erik Demaine z MIT: http://courses.csail.mit.edu/6.851/spring12/lectures/
- Textbook Brass 2008 Advanced data structures
Bloom Filter (Bloom 1970)
- supports insert x, test if x is in the set
- may give false positives, e.g. claim that x is in the set when it is not
- false negatives do not occur
Algorithm
- a bit string B[0,...,m-1] of length m, and k hash functions hi : U -> {0, ..., m-1}.
- insert(x): set bits B[h1(x)], ..., B[hk(x)] to 1.
- test if x in the set: compute h1(y), ..., hk(y) and check whether all these bits are 1
- if yes, claim x is in the set, but possibility of error
- if no, answer no, surely true
Bounding the error If hash functions are totally random and independent, the probability of error is approximately (1-exp(-nk/m))^k
- in fact probability somewhat higher for smaller values
- proof of the bound later
- totally random hash functions impractical (need to store hash value for each element of the universe), but the assumption simplifies analysis
- if we set k = ln(2) m/n, get error
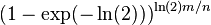 .
.
-
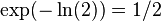 and thus we get error
and thus we get error 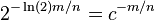 , where
, where 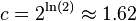
- to get error rate p for some n, we need
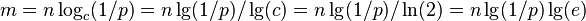
- for 1% error, we need about m=10n bits of space and k=7
- memory size and error rate are independent of the size of the universe U
- compare to a hash table, which needs at least to store data items themselves (e.g. in n lg u bits)
- if we used k=1 (Bloom filter with one hash function), we need m=n/ln(1/(1-p)) bits, which for p=0.01 is about 99.5n, about 10 times more than with 7 hash functions
Use of Bloom filters
- e.g. an approximate index of a larger data structure on a disk - if x not in the filter, do not bother looking at the disk, but small number of false positives not a problem
- Example: Google BigTable maps row label, column label and timestamp to a record, underlies many Google technologies. It uses Bloom filter to check if a combination of row and column label occur in a given fragment of the table
- For details, see Chang, Fay, et al. "Bigtable: A distributed storage system for structured data." ACM Transactions on Computer Systems (TOCS) 26.2 (2008): 4. [1]
- see also A. Z. Broder and M. Mitzenmacher. Network applications of Bloom filters: A survey. In Internet Math. Volume 1, Number 4 (2003), 485-509. [2]
Proof of the bound
- probability that some B[i] is set to 1 by hj(x) is 1/m
- probability that B[i] is not set to 1 is therefore 1-1/m and since we use k independent hash functions, the probability that B[i] is not set to one by any of them is (1-1/m)^k
- if we insert n elements, each is hashed by each function independently of other elements (hash functions are random) and thus Pr(B[i]=0)=(1-1/m)^{nk}
- Pr(B[i]=1) = 1-Pr(B[i]=0)
- consider a new element y which is not in the set
- error occurs when B[hj(y)]==1 for all j=1..k
- assume for now that these events are independent for different j
- error then occurs with probability Pr(B[i]=1)^k = (1-(1-1/m)^{nk})^k
- recall that
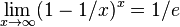
- thus probability of error is approximately (1-exp(-nk/m))^k
However, Pr(B[i]=1) are not independent
- consider the following experiment: have 2 binary random variables Y1, Y2, each generated independently, uniformly (i.e. P(Y1=1) = 0.5)
- Let X1 = Ya and X2 = Yb where a and b are chosen independently uniformly from set {1,2}
- X1 and X2 are not independent, e.g. Pr(X2 = 1|X1 = 1) = 2/3, whereas Pr(X2=1) = 1/2. (check the constants!)
- note: X1 and X2 would be independent conditioned on the sum of Y1+Y2.
- In Mitzenmacher & Upfal (2005), pp. 109–111, 308. - less strict independence assumption
Exercise Let us assume that we have separate Bloom filters for sets A and B with the same set k hash functions. How can we create Bloom filters for union and intersection? Will the result be different from filter created directly for the union or for the intersection?
Theory
- Bloom filter above use about 1.44 n lg(1/p) bits to achieve error rate p. There is lower bound of n lg(1/p) [Carter et al 1978], constant 1.44 can be improved to 1+o(1) using more complex data structures, which are then probably less practical
- Pagh, Anna, Rasmus Pagh, and S. Srinivasa Rao. "An optimal Bloom filter replacement." SODA 2005. [3]
- L. Carter, R. Floyd, J. Gill, G. Markowsky, and M. Wegman. Exact and approximate membership testers. STOC 1978, pages 59–65. [4]
- see also proof of lower bound in Broder and Mitzenmacher 2003 below
Counting Bloom filters
- support insert and delete
- each B[i] is a counter with b bits
- insert increases counters, delete decreases
- assume no overflows, or reserve largest value 2^b-1 as infinity, cannot be increased or decreased
- Fan, Li, et al. "Summary cache: A scalable wide-area web cache sharing protocol." ACM SIGCOMM Computer Communication Review. Vol. 28. No. 4. ACM, 1998. [5]
References
- Bloom, Burton H. (1970), "Space/Time Trade-offs in Hash Coding with Allowable Errors", Communications of the ACM 13 (7): 422–426
- Bloom filter. In Wikipedia, The Free Encyclopedia. Retrieved January 17, 2015, from http://en.wikipedia.org/w/index.php?title=Bloom_filter&oldid=641921171
- A. Z. Broder and M. Mitzenmacher. Network applications of Bloom filters: A survey. In Internet Math. Volume 1, Number 4 (2003), 485-509. [6]
Locality Sensitive Hashing
- todo: expand and according to slides
- typical hashing tables used for exact search: is x in set X?
- nearest neighbor(x): find y in set X minimizing d(x,y) for a given distance measure d
- or given x and distance threshold r, find all y in X with d(x,y)<=r
- want: polynomial time preprocessing of X, polylogarithmic queries
- later in the course: geometric data structures for similar problems in R^2 or higher dimensions
- but time or space grows exponentially with dimension (alternative is linear search) - "curse of dimensionality"
- examples of high-dimensional objects: images, documents, etc.
- locality sensitive hashing (LHS) is a randomized data structure not guaranteed to find the best answer but better running time
Approximate near neighbour (r,c)-NN
- preprocess a set X of size n for given r,c
- query(x):
- if there is y in X such that d(y,x)<=r, return z in X such that d(z,x)<=c*r
- LHS returns such z with some probability at least f
Example for Hamming distance in binary strings of length d
- Hash each string using L hashing functions, each using O(log n) randomly chosen string positions (hidden constant depends on d, r, c), then rehash using some other hash function to tables of size O(n) or a single table of size O(nL)
- Given q, find it using all L hash function, check every collision, report if distance at most cr. If checked more than 3L collisions, stop.
- L = O(n^{1/c})
- Space O(nL + nd), query time O(Ld+L(d/r)log n)
- Probability of failure less than some constant f < 1, can be boosted by repeating independently
Minimum hashing
- similarity of two sets measured as Jaccard index J(A,B) = |A intersect B | / | A union B |
- distance d(A,B) = 1-J(A,B)
- used e.g. for approximate duplicate document detection based on set of words occurring in the document
- choose a totally random hash function h
- minhash_h(A) = \min_{x\in A} h(x)
- Pr(minhash_h(A)=minhash_h(B)) = J(A,B) = 1-d(A,B) if using totally random hash function an no collisions in A union B
- where can be minimum in union? if in intersection, we get equality, otherwise not
- (r_1, r_2, 1-r_1, 1-r_2)-sensitive family (if we assume no collisions)
- again can we apply amplification
Notes
- Real-valued vectors: map to bits using random projections sgn (w · x + b)
- Data-dependent hashing: Use PCA, optimization, kernel tricks, even neural networks to learn hash functions good for a particular set of points and distance measure (or learn even distance measure from examples). [7]
Sources
- Har-Peled, Sariel, Piotr Indyk, and Rajeev Motwani. "Approximate Nearest Neighbor: Towards Removing the Curse of Dimensionality." Theory of Computing 8.1 (2012): 321-350. [8]
- Indyk and Motwani: Approximate nearest neighbors: Towards removing the curse of dimensionality. In Proc. 30th STOC, pp. 604–613. ACM Press, 1998.
- http://www.mit.edu/~andoni/LSH/ Pointers to papers
- Wikipedia
- Several lectures:
- http://infolab.stanford.edu/~ullman/mining/2009/similarity3.pdf various distances, connection to minhashing
- https://users.soe.ucsc.edu/~niejiazhong/slides/kumar.pdf nic intro, more duplicate detection, NN at the end
- http://delab.csd.auth.gr/courses/c_mmdb/mmdb-2011-2012-lsh2.pdf also nice intro through Hamming
- Charikar, Moses S. "Similarity estimation techniques from rounding algorithms." STOC 2002 [9]
- simHash (cosine measure)