ExonHunter
ExonHunter is a comprehensive gene finder based on hidden Markov
models allowing use of variety of additional sources of information
(ESTs, proteins, genome-genome comparison, repeats, and more).
It is described in the following papers:
-
Brona Brejova, Daniel G. Brown, Ming Li, Tomas Vinar. ExonHunter: a
comprehensive approach to gene finding. Bioinformatics, 2005. ISMB
2005.
-
Brona Brejova, Tomas Vinar, Yangyi Chen, Shengyue Wang, Guoping
Zhao, Daniel G. Brown, Ming Li, Yan Zhou. Finding genes in Schistosoma
japonicum: annotating novel genomes with help of extrinsic evidence.
Nucleic Acids Research, 37(7):e52. April 2009.
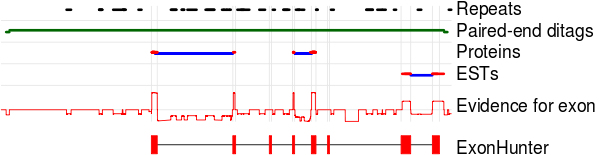
ExonHunter prediction on part of Schistosoma japonicum genome.
Download files:


